Compartmentalization of transcription and translation in Bacillus subtilis
- PMID: 10675340
- PMCID: PMC305609
- DOI: 10.1093/emboj/19.4.710
Compartmentalization of transcription and translation in Bacillus subtilis
Abstract
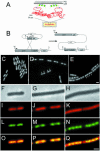
Using fusions of green fluorescent protein to subunits of RNA polymerase (RNAP) and ribosomes, we have investigated the subcellular localization of the transcriptional and translational machinery in the bacterium Bacillus subtilis. Unexpectedly, we found that RNAP resides principally within the nucleoid. Conversely, ribosomes localized almost exclusively outside the nucleoid, concentrating particularly towards sites of cell division. This zonal localization was not dependent on cell division and is probably due, at least in part, to exclusion from the nucleoid. Dual labelling of RNAP and ribosomes was used to confirm the spatial separation of the two processes. We conclude that, even in the absence of a nuclear membrane, transcription and translation occur predominantly in separate functional domains. At higher growth rates, concentrations of RNAP developed, probably representing the sites of rRNA synthesis. These may represent a further spatial specialization, possibly equivalent to the eukaryotic nucleolus.
Figures





References
-
- Beall B. and Lutkenhaus, J. (1991) FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev., 5, 447–455. - PubMed
-
- Bremer H. and Dennis,P.D. (1996) Modulation of chemical composition and other paramerters of the cell by growth rate. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Megasanik,B., Schaecter,M. and Umbarger.H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd edn. American Society for Microbiology Press, Washington, DC, pp. 1553–1569.
-
- Cashel M., Gentry,D.R., Hernandez,V.J. and Vinella,D. (1996) The stringent response. In Neidhardt,F.C., Ingraham,J.L., Low,K.B., Megasanik,B., Schaecter,M. and Umbarger.H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 2nd edn. American Society for Microbiology Press, Washington, DC, pp. 1458–1496.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

