The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway
- PMID: 10675346
- PMCID: PMC305615
- DOI: 10.1093/emboj/19.4.767
The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway
Abstract
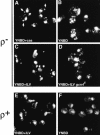
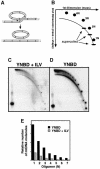
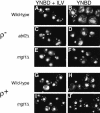
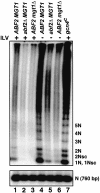
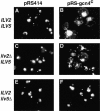
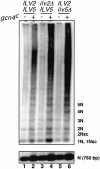
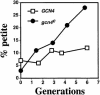
Mitochondrial DNA (mtDNA) is inherited as a protein-DNA complex (the nucleoid). We show that activation of the general amino acid response pathway in rho(+) and rho(-) petite cells results in an increased number of nucleoids without an increase in mtDNA copy number. In rho(-) cells, activation of the general amino acid response pathway results in increased intramolecular recombination between tandemly repeated sequences of rho(-) mtDNA to produce small, circular oligomers that are packaged into individual nucleoids, resulting in an approximately 10-fold increase in nucleoid number. The parsing of mtDNA into nucleoids due to general amino acid control requires Ilv5p, a mitochondrial protein that also functions in branched chain amino acid biosynthesis, and one or more factors required for mtDNA recombination. Two additional proteins known to function in mtDNA recombination, Abf2p and Mgt1p, are also required for parsing mtDNA into a larger number of nucleoids, although expression of these proteins is not under general amino acid control. Increased nucleoid number leads to increased mtDNA transmission, suggesting a mechanism to enhance mtDNA inheritance under amino acid starvation conditions.
Figures
References
-
- Bendich A.J. (1996) Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol., 255, 564–588. - PubMed
-
- Birky C.W., Strausberg, R.L., Perlman, P.S. and Forster, J.L. (1978) Vegetative segregation in yeast: estimating parameters using a random model. Mol. Gen. Genet., 158, 251–261.
-
- Brewer B.J. and Fangman, W.L. (1991) Mapping replication origins in yeast chromosomes. BioEssays, 13, 17–22. - PubMed
-
- Chen D.-C., Yang, B.-C. and Kuo, T.-T. (1992) One-step transformation of yeast in stationary phase. Curr. Genet., 21, 83–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

