Amino acid transporters are localized to transfer cells of developing pea seeds
- PMID: 10677425
- PMCID: PMC58869
- DOI: 10.1104/pp.122.2.319
Amino acid transporters are localized to transfer cells of developing pea seeds
Abstract
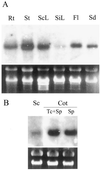
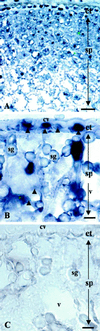
To determine the nature and cellular localization of amino acid transport in pea seeds, two cDNA clones belonging to the AAP family of H(+)/amino acid co-transporters (PsAAP1 and PsAAP2) were isolated from a cotyledon cDNA library of pea (Pisum sativum L.). Functional expression in the yeast amino acid uptake mutants 22Delta6AAL and 22Delta8AA showed that PsAAP1 mediates transport of neutral, acidic, and basic amino acids. RNA-blot analyses showed that PsAAP1 is expressed in seeds and vegetative organs, including amino acid sinks and sources, whereas PsAAP2 could not be detected. For developing seeds, transcripts of PsAAP1 were detected in coats and cotyledons, with seed coats giving a weak signal. In cotyledons, expression was highest in epidermal-transfer-cell-enriched tissue. RNA in situ hybridization analysis showed that PsAAP1 was predominantly present in epidermal transfer cells forming the outer surface of cotyledons, which abuts the seed coats. Overall, our observations suggest that this transporter, which is localized in transfer cells of cotyledons, might play a role in the uptake of the full spectrum of amino acids released from seed coats.
Figures
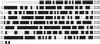

References
-
- Bick JA, Neelam A, Hall JL, Williams LE. Amino acid carriers of Ricinus communis expressed during seedling development: molecular cloning and expression analysis of two putative amino acid transporters, RcAAP1 and RcAAP2. Plant Mol Biol. 1998;36:377–385. - PubMed
-
- Borstlap AC, Schuurmans J. Kinetics of l-valine uptake in tobacco leaf discs: comparison of wild type, the digenic mutant Valr-2 and its monogenic derivatives. Planta. 1988;176:42–50. - PubMed
-
- De Jong A, Wolswinkel P. Differences in release of endogenous sugars and amino acids from attached and detached seed coats of developing pea seeds. Physiol Plant. 1995;94:78–86.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

