Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b(6)/f complex
- PMID: 10677443
- PMCID: PMC58887
- DOI: 10.1104/pp.122.2.505
Redox control of psbA gene expression in the cyanobacterium Synechocystis PCC 6803. Involvement of the cytochrome b(6)/f complex
Abstract
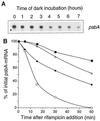
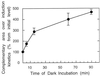
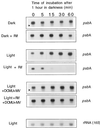
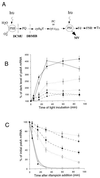
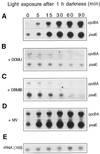
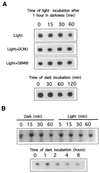
We investigated the role of the redox state of the photosynthetic and respiratory electron transport chains on the regulation of psbA expression in Synechocystis PCC 6803. Different means to modify the redox state of the electron carriers were used: (a) dark to oxidize the whole electron transport chain; (b) a shift from dark to light to induce its reduction; (c) the chemical interruption of the electron flow at different points to change the redox state of specific electron carriers; and (d) the presence of glucose to maintain a high reducing power in darkness. We show that changes in the redox state of the intersystem electron transport chain induce modifications of psbA transcript production and psbA mRNA stability. Reduction of the intersystem electron carriers activates psbA transcription and destabilizes the mRNA, while their oxidation induces a decrease in transcription and a stabilization of the transcript. Furthermore, our data suggest that the redox state of one of the electron carriers between the plastoquinone pool and photosystem I influences not only the expression of the psbA gene, but also that of other two photosynthetic genes, psaE and cpcBA. As a working hypothesis, we propose that the occupancy of the Q(0) site in the cytochrome b(6)/f complex may be involved in this regulation.
Figures



References
-
- Ajlani G, Kirilovsky D, Picaud M, Astier C. Molecular analysis of psbA mutations responsible for various herbicide resistance phenotypes in Synechocystis 6714. Plant Mol Biol. 1989;13:469–479. - PubMed
-
- Alfonso M, Perewoska I, Constant S, Kirilovsky D. Redox control of psbA expression in cyanobacterium Synechocystis strains. J Photochem Photobiol B Biol. 1999;48:104–113.
-
- Allen JF. Redox control of transcription: sensors, response regulators and repressors. FEBS Lett. 1993;323:203–207. - PubMed
-
- Aoki M, Katoh S. Size of the plastoquinone pool in photosynthetic and respiratory electron transport of Synechococcus sp. Plant Cell Physiol. 1983;24:1379–1386.
-
- Aro EM, Virgin I, Andersson B. Photoinhibition of PS II: inactivation, protein damage and turnover. Biochim Biophys Acta. 1993;1143:113–134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

