Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth
- PMID: 10677445
- PMCID: PMC58889
- DOI: 10.1104/pp.122.2.527
Auxin-regulated genes encoding cell wall-modifying proteins are expressed during early tomato fruit growth
Abstract
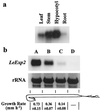
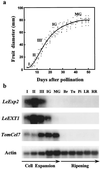
An expansin gene, LeExp2, was isolated from auxin-treated, etiolated tomato (Lycopersicon esculentum cv T5) hypocotyls. LeExp2 mRNA expression was restricted to the growing regions of the tomato hypocotyl and was up-regulated during incubation of hypocotyl segments with auxin. The pattern of expression of LeExp2 was also studied during tomato fruit growth, a developmental process involving rapid cell enlargement. The expression of genes encoding a xyloglucan endotransglycosylase (LeEXT1) and an endo-1, 4-beta-glucanase (Cel7), which, like LeExp2, are auxin-regulated in etiolated hypocotyls (C. Catalá, J.K.C. Rose, A.B. Bennett [1997] Plant J 12: 417-426), was also studied to examine the potential for synergistic action with expansins. LeExp2 and LeEXT1 genes were coordinately regulated, with their mRNA accumulation peaking during the stages of highest growth, while Cel7 mRNA abundance increased and remained constant during later stages of fruit growth. The expression of LeExp2, LeEXT1, and Cel7 was undetectable or negligible at the onset of and during fruit ripening, which is consistent with a specific role of these genes in regulating cell wall loosening during fruit growth, not in ripening-associated cell wall disassembly.
Figures



References
-
- Arrowsmith DA, de Silva J. Characterisation of two tomato fruit-expressed cDNAs encoding xyloglucan endo-transglycosylases. Plant Mol Biol. 1995;28:391–403. - PubMed
-
- Bonghi C, Ferrarese L, Ruperti B, Tonutti P, Ramina A. Endo-β-1,4-glucanases are involved in peach fruit growth and ripening, and regulated by ethylene. Physiol Plant. 1998;102:346–352.
-
- Brummell DA, Harpster MH, Dunsmuir P. Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol. 1999;39:161–169. - PubMed
-
- Brummell DA, Lashbrook CC, Bennett AB. Plant endo-1,4-β-glucanases: structure, properties and physiological function. Am Chem Soc Symp Ser. 1994;566:100–129.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

