Induction of an extracellular cyclic nucleotide phosphodiesterase as an accessory ribonucleolytic activity during phosphate starvation of cultured tomato cells
- PMID: 10677447
- PMCID: PMC58891
- DOI: 10.1104/pp.122.2.543
Induction of an extracellular cyclic nucleotide phosphodiesterase as an accessory ribonucleolytic activity during phosphate starvation of cultured tomato cells
Abstract
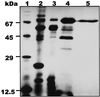
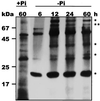
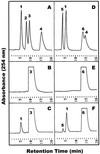
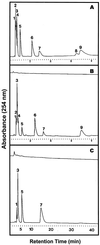
During growth under conditions of phosphate limitation, suspension-cultured cells of tomato (Lycopersicon esculentum Mill.) secrete phosphodiesterase activity in a similar fashion to phosphate starvation-inducible ribonuclease (RNase LE), a cyclizing endoribonuclease that generates 2':3'-cyclic nucleoside monophosphates (NMP) as its major monomeric products (T. Nürnberger, S. Abel, W. Jost, K. Glund [1990] Plant Physiol 92: 970-976). Tomato extracellular phosphodiesterase was purified to homogeneity from the spent culture medium of phosphate-starved cells and was characterized as a cyclic nucleotide phosphodiesterase. The purified enzyme has a molecular mass of 70 kD, a pH optimum of 6.2, and an isoelectric point of 8.1. The phosphodiesterase preparation is free of any detectable deoxyribonuclease, ribonuclease, and nucleotidase activity. Tomato extracellular phosphodiesterase is insensitive to EDTA and hydrolyzes with no apparent base specificity 2':3'-cyclic NMP to 3'-NMP and the 3':5'-cyclic isomers to a mixture of 3'-NMP and 5'-NMP. Specific activities of the enzyme are 2-fold higher for 2':3'-cyclic NMP than for 3':5'-cyclic isomers. Analysis of monomeric products of sequential RNA hydrolysis with purified RNase LE, purified extracellular phosphodiesterase, and cleared -Pi culture medium as a source of 3'-nucleotidase activity indicates that cyclic nucleotide phosphodiesterase functions as an accessory ribonucleolytic activity that effectively hydrolyzes primary products of RNase LE to substrates for phosphate-starvation-inducible phosphomonoesterases. Biosynthetical labeling of cyclic nucleotide phopshodiesterase upon phosphate starvation suggests de novo synthesis and secretion of a set of nucleolytic enzymes for scavenging phosphate from extracellular RNA substrates.
Figures


References
-
- Abel S, Glund K. Ribonuclease in plant vacuoles: purification and molecular properties of the enzyme from cultured tomato cells. Planta. 1987;172:71–78. - PubMed
-
- Abel S, Krauss G-J, Glund K. Ribonuclease in tomato vacuoles: high-performance liquid chromatographic analysis of ribonucleolytic activities and base specificity. Biochim Biophys Acta. 1989;998:145–150.
-
- Bieleski RL. Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol. 1973;24:225–252.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

