Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I
- PMID: 10677490
- PMCID: PMC26463
- DOI: 10.1073/pnas.040549897
Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I
Abstract
Current evidence suggests that the length of poly(A) tails of bacterial mRNAs result from a competition between poly(A) polymerase and exoribonucleases that attack the 3' ends of RNAs. Here, we show that host factor Hfq is also involved in poly(A) tail metabolism. Inactivation of the hfq gene reduces the length of poly(A) tails synthesized at the 3' end of the rpsO mRNA by poly(A) polymerase I in vivo. In vitro, Hfq stimulates synthesis of long tails by poly(A) polymerase I. The strong binding of Hfq to oligoadenylated RNA probably explains why it stimulates elongation of primers that already harbor tails of 20-35 A. Polyadenylation becomes processive in the presence of Hfq. The similar properties of Hfq and the PABPII poly(A) binding protein, which stimulates poly(A) tail elongation in mammals, indicates that similar mechanisms control poly(A) tail synthesis in prokaryotes and eukaryotes.
Figures


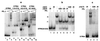

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

