Transition-state structure as a unifying basis in protein-folding mechanisms: contact order, chain topology, stability, and the extended nucleus mechanism
- PMID: 10677494
- PMCID: PMC26468
- DOI: 10.1073/pnas.97.4.1525
Transition-state structure as a unifying basis in protein-folding mechanisms: contact order, chain topology, stability, and the extended nucleus mechanism
Abstract
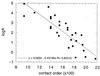
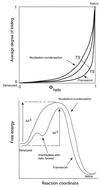
I attempt to reconcile apparently conflicting factors and mechanisms that have been proposed to determine the rate constant for two-state folding of small proteins, on the basis of general features of the structures of transition states. Phi-Value analysis implies a transition state for folding that resembles an expanded and distorted native structure, which is built around an extended nucleus. The nucleus is composed predominantly of elements of partly or well-formed native secondary structure that are stabilized by local and long-range tertiary interactions. These long-range interactions give rise to connecting loops, frequently containing the native loops that are poorly structured. I derive an equation that relates differences in the contact order of a protein to changes in the length of linking loops, which, in turn, is directly related to the unfavorable free energy of the loops in the transition state. Kinetic data on loop extension mutants of CI2 and alpha-spectrin SH3 domain fit the equation qualitatively. The rate of folding depends primarily on the interactions that directly stabilize the nucleus, especially those in native-like secondary structure and those resulting from the entropy loss from the connecting loops, which vary with contact order. This partitioning of energy accounts for the success of some algorithms that predict folding rates, because they use these principles either explicitly or implicitly. The extended nucleus model thus unifies the observations of rate depending on both stability and topology.
Figures


References
-
- Plaxco K W, Simons K T, Baker D. J Mol Biol. 1998;277:985–994. - PubMed
-
- Matouschek A, Kellis J T, Jr, Serrano L, Fersht A R. Nature (London) 1989;340:122–126. - PubMed
-
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1999.
-
- Fersht A R, Leatherbarrow R J, Wells T N C. Nature (London) 1986;322:284–286.
-
- Fersht A R, Leatherbarrow R, Wells T N C. Biochemistry. 1987;26:6030–6038. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

