A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase
- PMID: 10677495
- PMCID: PMC26469
- DOI: 10.1073/pnas.030539597
A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase
Abstract
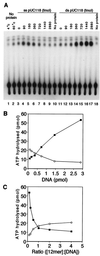
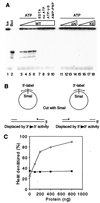
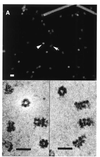
The minichromosome maintenance (MCM) proteins are essential for DNA replication in eukaryotes. Thus far, all eukaryotes have been shown to contain six highly related MCMs that apparently function together in DNA replication. Sequencing of the entire genome of the thermophilic archaeon Methanobacterium thermoautotrophicum has allowed us to identify only a single MCM-like gene (ORF Mt1770). This gene is most similar to MCM4 in eukaryotic cells. Here we have expressed and purified the M. thermoautotrophicum MCM protein. The purified protein forms a complex that has a molecular mass of approximately 850 kDa, consistent with formation of a double hexamer. The protein has an ATP-independent DNA-binding activity, a DNA-stimulated ATPase activity that discriminates between single- and double-stranded DNA, and a strand-displacement (helicase) activity that can unwind up to 500 base pairs. The 3' to 5' helicase activity requires both ATP hydrolysis and a functional nucleotide-binding site. Moreover, the double hexamer form is the active helicase. It is therefore likely that an MCM complex acts as the replicative DNA helicase in eukaryotes and archaea. The simplified replication machinery in archaea may provide a simplified model for assembly of the machinery required for initiation of eukaryotic DNA replication.
Figures



References
-
- Tye B K. Annu Rev Biochem. 1999;68:649–686. - PubMed
-
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Cell. 1995;81:601–609. - PubMed
-
- Chong J P, Mahbubani H M, Khoo C Y, Blow J J. Nature (London) 1995;375:418–421. - PubMed
-
- Madine M A, Khoo C Y, Mills A D, Mushal C, Laskey R A. Curr Biol. 1995;5:1270–1279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

