Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs
- PMID: 10679009
- PMCID: PMC14788
- DOI: 10.1091/mbc.11.2.497
Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs
Abstract
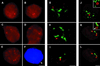
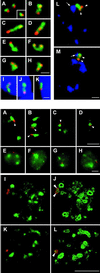
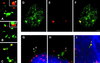
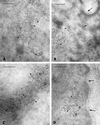
In the present study, the spatial organization of intron-containing pre-mRNAs of Epstein-Barr virus (EBV) genes relative to location of splicing factors is investigated. The intranuclear position of transcriptionally active EBV genes, as well as of nascent transcripts, is found to be random with respect to the speckled accumulations of splicing factors (SC35 domains) in Namalwa cells, arguing against the concept of the locus-specific organization of mRNA genes with respect to the speckles. Microclusters of splicing factors are, however, frequently superimposed on nascent transcript sites. The transcript environment is a dynamic structure consisting of both nascent and released transcripts, i.e., the track-like transcript environment. Both EBV sequences of the chromosome 1 homologue are usually associated with the track, are transcriptionally active, and exhibit in most cases a polar orientation. In contrast to nascent transcripts (in the form of spots), the association of a post-transcriptional pool of viral pre-mRNA (in the form of tracks) with speckles is not random and is further enhanced in transcriptionally silent cells when splicing factors are sequestered in enlarged accumulations. The transcript environment reflects the intranuclear transport of RNA from the sites of transcription to SC35 domains, as shown by concomitant mapping of DNA, RNA, and splicing factors. No clear vectorial intranuclear trafficking of transcripts from the site of synthesis toward the nuclear envelope for export into the cytoplasm is observed. Using Namalwa and Raji cell lines, a correlation between the level of viral gene transcription and splicing factor accumulation within the viral transcript environment has been observed. This supports a concept that the level of transcription can alter the spatial relationship among intron-containing genes, their transcripts, and speckles attributable to various levels of splicing factors recruited from splicing factor reservoirs. Electron microscopic in situ hybridization studies reveal that the released transcripts are directed toward reservoirs of splicing factors organized in clusters of interchromatin granules. Our results point to the bidirectional intranuclear movement of macromolecular complexes between intron-containing genes and splicing factor reservoirs: the recruitment of splicing factors to transcription sites and movement of released transcripts from DNA loci to reservoirs of splicing factors.
Figures

References
-
- Aspegren A, Rabino C, Bridge E. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp Cell Res. 1998;245:203–213. - PubMed
-
- Baurén G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. - PubMed
-
- Bendayan M, Nanci A, Kan FWK. Effect of tissue processing on colloidal gold cytochemistry. J Histochem Cytochem. 1987;35:983–996. - PubMed
-
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

