Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast
- PMID: 10679030
- PMCID: PMC14809
- DOI: 10.1091/mbc.11.2.773
Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast
Abstract
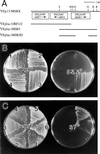
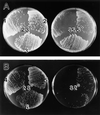
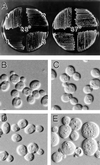
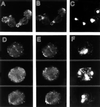
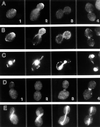
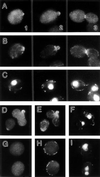
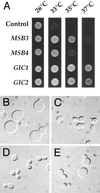
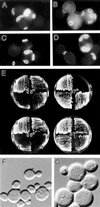
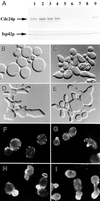
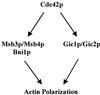
In the yeast Saccharomyces cerevisiae, Cdc24p functions at least in part as a guanine-nucleotide-exchange factor for the Rho-family GTPase Cdc42p. A genetic screen designed to identify possible additional targets of Cdc24p instead identified two previously known genes, MSB1 and CLA4, and one novel gene, designated MSB3, all of which appear to function in the Cdc24p-Cdc42p pathway. Nonetheless, genetic evidence suggests that Cdc24p may have a function that is distinct from its Cdc42p guanine-nucleotide-exchange factor activity; in particular, overexpression of CDC42 in combination with MSB1 or a truncated CLA4 in cells depleted for Cdc24p allowed polarization of the actin cytoskeleton and polarized cell growth, but not successful cell proliferation. MSB3 has a close homologue (designated MSB4) and two more distant homologues (MDR1 and YPL249C) in S. cerevisiae and also has homologues in Schizosaccharomyces pombe, Drosophila (pollux), and humans (the oncogene tre17). Deletion of either MSB3 or MSB4 alone did not produce any obvious phenotype, and the msb3 msb4 double mutant was viable. However, the double mutant grew slowly and had a partial disorganization of the actin cytoskeleton, but not of the septins, in a fraction of cells that were larger and rounder than normal. Like Cdc42p, both Msb3p and Msb4p localized to the presumptive bud site, the bud tip, and the mother-bud neck, and this localization was Cdc42p dependent. Taken together, the data suggest that Msb3p and Msb4p may function redundantly downstream of Cdc42p, specifically in a pathway leading to actin organization. From previous work, the BNI1, GIC1, and GIC2 gene products also appear to be involved in linking Cdc42p to the actin cytoskeleton. Synthetic lethality and multicopy suppression analyses among these genes, MSB, and MSB4, suggest that the linkage is accomplished by two parallel pathways, one involving Msb3p, Msb4p, and Bni1p, and the other involving Gic1p and Gic2p. The former pathway appears to be more important in diploids and at low temperatures, whereas the latter pathway appears to be more important in haploids and at high temperatures.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

