Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain
- PMID: 10680765
- PMCID: PMC6871833
- DOI: 10.1002/(sici)1097-0193(200002)9:2<81::aid-hbm3>3.0.co;2-8
Mathematical/computational challenges in creating deformable and probabilistic atlases of the human brain
Abstract
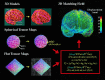
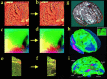
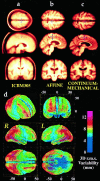
Striking variations in brain structure, especially in the gyral patterns of the human cortex, present fundamental challenges in human brain mapping. Probabilistic brain atlases, which encode information on structural and functional variability in large human populations, are powerful research tools with broad applications. Knowledge-based imaging algorithms can also leverage atlased information on anatomic variation. Applications include automated image labeling, pathology detection in individuals or groups, and investigating how regional anatomy is altered in disease, and with age, gender, handedness and other clinical or genetic factors. In this report, we illustrate some of the mathematical challenges involved in constructing population-based brain atlases. A disease-specific atlas is constructed to represent the human brain in Alzheimer's disease (AD). Specialized strategies are developed for population-based averaging of anatomy. Sets of high-dimensional elastic mappings, based on the principles of continuum mechanics, reconfigure the anatomy of a large number of subjects in an anatomic image database. These mappings generate a local encoding of anatomic variability and are used to create a crisp anatomical image template with highly resolved structures in their mean spatial location. Specialized approaches are also developed to average cortical topography. Since cortical patterns are altered in a variety of diseases, gyral pattern matching is used to encode the magnitude and principal directions of local cortical variation. In the resulting cortical templates, subtle features emerge. Regional asymmetries appear that are not apparent in individual anatomies. Population-based maps of cortical variation reveal a mosaic of variability patterns that segregate sharply according to functional specialization and cytoarchitectonic boundaries.
Figures
Similar articles
-
Disease-Specific Probabilistic Brain Atlases.Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2000 Jun 11;2000:227-234. doi: 10.1109/MMBIA.2000.852382. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2000. PMID: 19424457 Free PMC article.
-
Detection and mapping of abnormal brain structure with a probabilistic atlas of cortical surfaces.J Comput Assist Tomogr. 1997 Jul-Aug;21(4):567-81. doi: 10.1097/00004728-199707000-00008. J Comput Assist Tomogr. 1997. PMID: 9216760
-
Detection, visualization and animation of abnormal anatomic structure with a deformable probabilistic brain atlas based on random vector field transformations.Med Image Anal. 1997 Sep;1(4):271-94. doi: 10.1016/s1361-8415(97)85002-5. Med Image Anal. 1997. PMID: 9873911
-
Maps of the brain.Anat Rec. 2001 Apr;265(2):37-53. doi: 10.1002/ar.1057. Anat Rec. 2001. PMID: 11323769 Review.
-
Probabilistic approaches for atlasing normal and disease-specific brain variability.Anat Embryol (Berl). 2001 Oct;204(4):267-82. doi: 10.1007/s004290100198. Anat Embryol (Berl). 2001. PMID: 11720233 Review.
Cited by
-
Hippocampal shape and volume changes with antipsychotics in early stage psychotic illness.Front Psychiatry. 2012 Nov 12;3:96. doi: 10.3389/fpsyt.2012.00096. eCollection 2012. Front Psychiatry. 2012. PMID: 23162479 Free PMC article.
-
A system for the generation of curves on 3D brain images.Hum Brain Mapp. 2001 Sep;14(1):1-15. doi: 10.1002/hbm.1037. Hum Brain Mapp. 2001. PMID: 11500986 Free PMC article.
-
Brain image registration using cortically constrained harmonic mappings.Inf Process Med Imaging. 2007;20:359-71. doi: 10.1007/978-3-540-73273-0_30. Inf Process Med Imaging. 2007. PMID: 17633713 Free PMC article.
-
Cortical surface segmentation and mapping.Neuroimage. 2004;23 Suppl 1(0 1):S108-18. doi: 10.1016/j.neuroimage.2004.07.042. Neuroimage. 2004. PMID: 15501080 Free PMC article. Review.
-
Hierarchical unbiased graph shrinkage (HUGS): a novel groupwise registration for large data set.Neuroimage. 2014 Jan 1;84:626-38. doi: 10.1016/j.neuroimage.2013.09.023. Epub 2013 Sep 19. Neuroimage. 2014. PMID: 24055505 Free PMC article.
References
-
- Ashburner J, Friston K. 1997. Multimodal image coregistration and partitioning — a unified framework. NeuroImage 6:209–217. - PubMed
-
- Bookstein FL. 1997. Landmark methods for forms without landmarks: morphometrics of group differences in outline shape. Med Image Analysis 1: 225–243. - PubMed
-
- Cao J, Worsley KJ. 1999. The geometry of the Hotelling's T‐squared random field with applications to the detection of shape changes. Annals of Statistics (in press).
-
- Christensen GE, Rabbitt RD, Miller MI. 1993. A deformable neuroanatomy textbook based on viscous fluid mechanics. 27th Ann. Conf. on Inf. Sciences and Systems, p 211–216.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

