Toward Anopheles transformation: Minos element activity in anopheline cells and embryos
- PMID: 10681436
- PMCID: PMC15770
- DOI: 10.1073/pnas.040568397
Toward Anopheles transformation: Minos element activity in anopheline cells and embryos
Erratum in
- Proc Natl Acad Sci U S A 2000 May 23;97(11):6236
Abstract
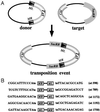
The ability of the Minos transposable element to function as a transformation vector in anopheline mosquitoes was assessed. Two recently established Anopheles gambiae cell lines were stably transformed by using marked Minos transposons in the presence of a helper plasmid expressing transposase. The markers were either the green fluorescent protein or the hygromycin B phosphotransferase gene driven by the Drosophila Hsp70 promoter. Cloning and sequencing of the integration sites demonstrated that insertions in the cell genome occurred through the action of Minos transposase. Furthermore, an interplasmid transposition assay established that Minos transposase is active in the cytoplasmic environment of Anopheles stephensi embryos: interplasmid transposition events isolated from injected preblastoderm embryos were identified as Minos transposase-mediated integrations, and no events were recorded in the absence of an active transposase. These results demonstrate that Minos vectors are suitable candidates for germ-line transformation of anopheline mosquitoes.
Figures

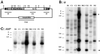


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

