The accuracy of codon recognition by polypeptide release factors
- PMID: 10681447
- PMCID: PMC15751
- DOI: 10.1073/pnas.030541097
The accuracy of codon recognition by polypeptide release factors
Abstract
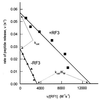
The precision with which individual termination codons in mRNA are recognized by protein release factors (RFs) has been measured and compared with the decoding of sense codons by tRNA. An Escherichia coli system for protein synthesis in vitro with purified components was used to study the accuracy of termination by RF1 and RF2 in the presence or absence of RF3. The efficiency of factor-dependent termination at all sense codons differing from any of the three stop codons by a single mutation was measured and compared with the efficiency of termination at the three stop codons. RF1 and RF2 discriminate against sense codons related to stop codons by between 3 and more than 6 orders of magnitude. This high level of accuracy is obtained without energy-driven error correction (proofreading), in contrast to codon-dependent aminoacyl-tRNA recognition by ribosomes. Two codons, UAU and UGG, stand out as hotspots for RF-dependent premature termination.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

