Spatial separation of parental genomes in preimplantation mouse embryos
- PMID: 10684246
- PMCID: PMC2169371
- DOI: 10.1083/jcb.148.4.629
Spatial separation of parental genomes in preimplantation mouse embryos
Abstract
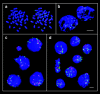
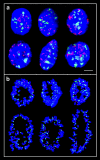
We have used two different experimental approaches to demonstrate topological separation of parental genomes in preimplantation mouse embryos: mouse eggs fertilized with 5-bromodeoxyuridine (BrdU)-labeled sperm followed by detection of BrdU in early diploid embryos, and differential heterochromatin staining in mouse interspecific hybrid embryos. Separation of chromatin according to parental origin was preserved up to the four-cell embryo stage and then gradually disappeared. In F1 hybrid animals, genome separation was also observed in a proportion of somatic cells. Separate nuclear compartments during preimplantation development, when extreme chromatin remodelling occurs, and possibly in some differentiated cell types, may be associated with epigenetic reprogramming.
Figures

Similar articles
-
The battle of the sexes after fertilization: behaviour of paternal and maternal chromosomes in the early mammalian embryo.Chromosome Res. 2001;9(4):263-71. doi: 10.1023/a:1016686312142. Chromosome Res. 2001. PMID: 11419791 Review.
-
3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development.BMC Dev Biol. 2012 Oct 24;12:30. doi: 10.1186/1471-213X-12-30. BMC Dev Biol. 2012. PMID: 23095683 Free PMC article.
-
H3S10 phosphorylation marks constitutive heterochromatin during interphase in early mouse embryos until the 4-cell stage.J Reprod Dev. 2012;58(4):467-75. doi: 10.1262/jrd.11-109h. Epub 2012 Apr 27. J Reprod Dev. 2012. PMID: 22572731
-
Three-dimensional analysis of nuclear heterochromatin distribution during early development in the rabbit.Chromosoma. 2018 Sep;127(3):387-403. doi: 10.1007/s00412-018-0671-z. Epub 2018 Apr 18. Chromosoma. 2018. PMID: 29666907 Free PMC article.
-
Non-canonical spatial organization of heterochromatin in mouse preimplantation embryos.Reproduction. 2025 Jan 9;169(2):e240271. doi: 10.1530/REP-24-0271. Print 2025 Feb 1. Reproduction. 2025. PMID: 39555985 Review.
Cited by
-
Epigenetic reprogramming in the mammalian embryo: struggle of the clones.Genome Biol. 2002;3(2):REVIEWS1003. doi: 10.1186/gb-2002-3-2-reviews1003. Epub 2002 Jan 29. Genome Biol. 2002. PMID: 11864375 Free PMC article. Review.
-
Making sense out of syngamy at the onset of mammalian development.J Assist Reprod Genet. 2018 Aug;35(8):1357-1358. doi: 10.1007/s10815-018-1282-6. J Assist Reprod Genet. 2018. PMID: 30069851 Free PMC article. No abstract available.
-
The battle of the sexes after fertilization: behaviour of paternal and maternal chromosomes in the early mammalian embryo.Chromosome Res. 2001;9(4):263-71. doi: 10.1023/a:1016686312142. Chromosome Res. 2001. PMID: 11419791 Review.
-
Evidence for conserved DNA and histone H3 methylation reprogramming in mouse, bovine and rabbit zygotes.Epigenetics Chromatin. 2008 Nov 3;1(1):8. doi: 10.1186/1756-8935-1-8. Epigenetics Chromatin. 2008. PMID: 19014417 Free PMC article.
-
C. elegans pronuclei fuse after fertilization through a novel membrane structure.J Cell Biol. 2020 Feb 3;219(2):e201909137. doi: 10.1083/jcb.201909137. J Cell Biol. 2020. PMID: 31834351 Free PMC article.
References
-
- Adenot P.G., Mercier Y., Renard J.-P., Thompson E.M. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615–4625. - PubMed
-
- Aoki F., Worrad D.M., Schultz R.M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997;181:296–307. - PubMed
-
- Bouniol-Baly C., Nguyen E., Besombes D., Debey P. Dynamic organization of DNA replication in one-cell mouse embryosrelationship to transcriptional activation. Exp. Cell Res. 1997;236:201–211. - PubMed
-
- Cattanach B.M., Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. - PubMed
-
- Fundele R., Surani M.A. Experimental embryological analysis of genetic imprinting in mouse development. Dev. Genet. 1994;15:515–522. - PubMed

