Evolutionary rate and genetic drift of hepatitis C virus are not correlated with the host immune response: studies of infected donor-recipient clusters
- PMID: 10684268
- PMCID: PMC111742
- DOI: 10.1128/jvi.74.6.2541-2549.2000
Evolutionary rate and genetic drift of hepatitis C virus are not correlated with the host immune response: studies of infected donor-recipient clusters
Abstract
Six donor-recipient clusters of hepatitis C virus (HCV)-infected individuals were studied. For five clusters the period of infection of the donor could be estimated, and for all six clusters the time of infection of the recipients from the donor via blood transfusion was also precisely known. Detailed phylogenetic analyses were carried out to investigate the genomic evolution of the viral quasispecies within infected individuals in each cluster. The molecular clock analysis showed that HCV quasispecies within a patient are evolving at the same rate and that donors that have been infected for longer time tend to have a lower evolutionary rate. Phylogenetic analysis based on the split decomposition method revealed different evolutionary patterns in different donor-recipient clusters. Reactivity of antibody against the first hypervariable region (HVR1) of HCV in donor and recipient sera was evaluated and correlated to the calculated evolutionary rate. Results indicate that anti-HVR1 reactivity was related more to the overall level of humoral immune response of the host than to the HVR1 sequence itself, suggesting that the particular sequence of the HVR1 peptides is not the determinant of reactivity. Moreover, no correlation was found between the evolutionary rate or the heterogeneity of the viral quasispecies in the patients and the strength of the immune response to HVR1 epitopes. Rather, the results seem to imply that genetic drift is less dependent on immune pressure than on the rate of evolution and that the genetic drift of HCV is independent of the host immune pressure.
Figures
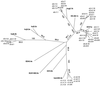
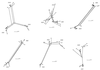


References
-
- Abe K, Inchauspé G, Fujisawa K. Genomic characterisation and mutation rate of hepatitis C virus isolated from a patient who contracted hepatitis during an epidemic on non-A, non-B hepatitis in Japan. J Gen Virol. 1992;72:2725–2729. - PubMed
-
- Aiyama T, Yoshioka K, Okumura A, Takayanagi M, Iwata K, Ishikawa T, Kakuma S. Sequence analysis of hypervariable region of hepatitis C virus (HCV) associated with immune complex in patients with chronic HCV infection. J Infect Dis. 1996;174:1316–1320. - PubMed
-
- Allain J-P, Zhai W, Shang D, Timmers E, Alexander G J M. Hypervariable region diversity of hepatitis C virus and humoral response: comparison between patients with and without cirrhosis. J Med Virol. 1999;59:25–31. - PubMed
-
- Casino C, McAllister J, Davidson F, Power J, Lawlord E, Yap P L, Simmonds P, Smith D B. Variation of hepatitis C virus following serial transmission: multiple mechanisms of diversification of the hypervariable region and evidence for convergent genome evolution. J Gen Virol. 1999;80:717–725. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

