Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting
- PMID: 10684292
- PMCID: PMC111766
- DOI: 10.1128/jvi.74.6.2760-2769.2000
Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting
Abstract
We present a detailed and quantitative analysis of the functional characteristics of the 1,000-nucleotide segment at the 5' end of the human immunodeficiency virus type 1 (HIV-1) RNA genome. This segment of the viral genome contains several important cis-acting sequences, including the TAR, polyadenylation, viral att site, minus-strand primer-binding site, and 5' splice donor sequences, as well as coding sequences for the matrix protein and the N-terminal half of the capsid protein. The genetic footprinting technique was used to determine quantitatively the abilities of 134 independent insertion mutations to (i) make stable viral RNA, (ii) assemble and release viral RNA-containing viral particles, and (iii) enter host cells, complete reverse transcription, enter the nuclei of host cells, and generate proviruses in the host genome by integration. All of the mutants were constructed and analyzed en masse, greatly decreasing the labor typically involved in mutagenesis studies. The results confirmed the presence of several previously known functional features in this region of the HIV genome and provided evidence for several novel features, including newly identified cis-acting sequences that appeared to contribute to (i) the formation of stable viral transcripts, (ii) viral RNA packaging, and (iii) an early step in viral replication. The results also pointed to an unanticipated trans-acting role for the N-terminal portion of matrix in the formation of stable viral RNA transcripts. Finally, in contrast to previous reports, the results of this study suggested that detrimental mutations in the matrix and capsid proteins principally interfered with viral assembly.
Figures
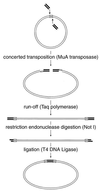
 ), the target DNA plasmid is represented by thin lines, and the 5-bp sequences in the target DNA that were duplicated during mutagenesis are represented by open boxes (
), the target DNA plasmid is represented by thin lines, and the 5-bp sequences in the target DNA that were duplicated during mutagenesis are represented by open boxes ( ).
).

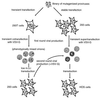
 ), replication-defective viral genome with mutation in trans-acting factor (
), replication-defective viral genome with mutation in trans-acting factor ( ), wild-type viral protein (
), wild-type viral protein ( ), and mutant viral protein (
), and mutant viral protein ( ).
).
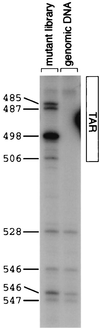
 ) and one biotinylated primer (
) and one biotinylated primer ( ). The PCR products are captured by streptavidin-agarose resin (
). The PCR products are captured by streptavidin-agarose resin ( ) and digested with a restriction enzyme that recognizes a site in the insertion sequence. The radioactively labeled ends of the PCR products are released.
) and digested with a restriction enzyme that recognizes a site in the insertion sequence. The radioactively labeled ends of the PCR products are released.

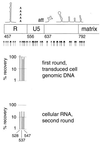
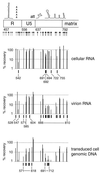

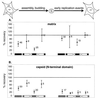
 [amino acids 1 to 132; nucleotides 792 to 1187]), the N-terminal region (
[amino acids 1 to 132; nucleotides 792 to 1187]), the N-terminal region ( [amino acids 1 to 35; nucleotides 792 to 896]), the central region (
[amino acids 1 to 35; nucleotides 792 to 896]), the central region ( [amino acids 36 to 102; nucleotides 897 to 1097]), and the C-terminal region (
[amino acids 36 to 102; nucleotides 897 to 1097]), and the C-terminal region ( [amino acids 104 to 130; nucleotides 1101 to 1181]). (B) Data for the N-terminal half of the capsid protein. Data are shown for insertions in the N-terminal half (
[amino acids 104 to 130; nucleotides 1101 to 1181]). (B) Data for the N-terminal half of the capsid protein. Data are shown for insertions in the N-terminal half ( [amino acids 1 to 101; nucleotides 1188 to 1490]), the N-terminal beta hairpin (
[amino acids 1 to 101; nucleotides 1188 to 1490]), the N-terminal beta hairpin ( [amino acids 1 to 15; nucleotides 1188 to 1232]), the central helical region (
[amino acids 1 to 15; nucleotides 1188 to 1232]), the central helical region ( [amino acids 17 to 81, nucleotides 1236 to 1430]), and the cyclophilin A-binding region (
[amino acids 17 to 81, nucleotides 1236 to 1430]), and the cyclophilin A-binding region ( [amino acids 85 to 96; nucleotides 1440 to 1475]).
[amino acids 85 to 96; nucleotides 1440 to 1475]).References
-
- Baker T A, Mizuuchi M, Savilahti H, Mizuuchi K. Division of labor among monomers within the Mu transposase tetramer. Cell. 1993;74:723–733. - PubMed
-
- Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. - PubMed
-
- Bukrinskaya A G, Vorkunova G K, Tentsov Y. HIV-1 matrix protein p17 resides in cell nuclei in association with genomic RNA. AIDS Res Hum Retroviruses. 1992;8:1795–1801. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

