Identification of a boundary domain adjacent to the potent human cytomegalovirus enhancer that represses transcription of the divergent UL127 promoter
- PMID: 10684299
- PMCID: PMC111773
- DOI: 10.1128/jvi.74.6.2826-2839.2000
Identification of a boundary domain adjacent to the potent human cytomegalovirus enhancer that represses transcription of the divergent UL127 promoter
Abstract
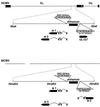
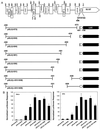
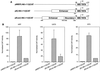
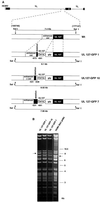
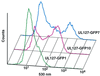
Transcriptional repression within a complex modular promoter may play a key role in determining the action of enhancer elements. In human cytomegalovirus, the major immediate-early promoter (MIEP) locus contains a highly potent and complex modular enhancer. Evidence is presented suggesting that sequences of the MIEP between nucleotide positions -556 and -673 function to prevent transcription activation by enhancer elements from the UL127 open reading frame divergent promoter. Transient transfection assays of reporter plasmids revealed repressor sequences located between nucleotides -556 and -638. The ability of these sequences to confer repression in the context of an infection was shown using recombinant viruses generated from a bacterial artificial chromosome containing an infectious human cytomegalovirus genome. In addition to repressor sequences between -556 and -638, infection experiments using recombinant virus mutants indicated that sequences between -638 and -673 also contribute to repression of the UL127 promoter. On the basis of in vitro transcription and transient transfection assays, we further show that interposed viral repressor sequences completely inhibit enhancer-mediated activation of not only the homologous but also heterologous promoters. These and other experiments suggest that repression involves an interaction of host-encoded regulatory factors with defined promoter sequences that have the property of proximally interfering with upstream enhancer elements in a chromatin-independent manner. Altogether, our findings establish the presence of a boundary domain that efficiently blocks enhancer-promoter interactions, thus explaining how the enhancer can work to selectively activate the MIEP.
Figures





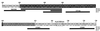
References
-
- Adhya S, Geanacopoulos M, Lewis D E, Roy S, Aki T. Transcription regulation by repressosome and by RNA polymerase contact. Cold Spring Harbor Symp Quant Biol. 1998;63:1–9. - PubMed
-
- Angulo A, Suto C, Heyman R A, Ghazal P. Characterization of the sequences of the human cytomegalovirus enhancer that mediate differential regulation by natural and synthetic retinoids. Mol Endocrinol. 1996;10:781–793. - PubMed
-
- Baskar J F, Smith P P, Nilaver G, Jupp R A, Hoffmann S, Peffer N J, Tenney D J, Colberg-Poley A M, Ghazal P, Nelson J. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell-type-specific expression in transgenic mice. J Virol. 1996;70:3207–3214. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

