dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex
- PMID: 10684876
- PMCID: PMC6772911
- DOI: 10.1523/JNEUROSCI.20-05-01746.2000
dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex
Abstract
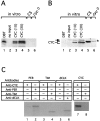
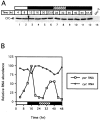
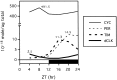
In Drosophila melanogaster four circadian clock proteins termed PERIOD (PER), TIMELESS (TIM), dCLOCK (dCLK), and CYCLE (CYC/dBMAL1) function in a transcriptional feedback loop that is a core element of the oscillator mechanism. dCLK and CYC are members of the basic helix-loop-helix (bHLH)/PAS (PER-ARNT-SIM) superfamily of transcription factors and are required for high-level expression of per and tim and repression of dClk, whereas PER and TIM inhibit dCLK-CYC-mediated transcription and lead to the activation of dClk. To understand further the dynamic regulation within the circadian oscillator mechanism, we biochemically characterized in vivo-produced CYC, determined the interactions of the four clock proteins, and calculated their absolute levels as a function of time. Our results indicate that throughout a daily cycle the majority of the dCLK present in adult heads stably interacts with CYC, indicating that CYC is the primary in vivo partner of dCLK. dCLK-CYC dimers are bound by PER and TIM during the late evening and early morning, suggesting the formation of a tetrameric complex with impaired transcriptional activity. Although dCLK is present in limiting amounts and CYC is by far the most abundant of the four clock proteins that have been examined, PER and TIM appear to interact preferentially with dCLK. Our results suggest that dCLK is the main component regulating the daily abundance of transcriptionally active dCLK-CYC complexes.
Figures


References
-
- Allada R, White NE, So WV, Hall JC, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
-
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Steve AK. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. - PubMed
-
- Citri Y, Colot HV, Jacquier AC, Yu Q, Hall JC, Baltimore D, Rosbash M. A family of unusually spliced biologically active transcripts encoded by a Drosophila clock gene. Nature. 1987;326:42–47. - PubMed
-
- Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
