Interaction of the yeast DExH-box RNA helicase prp22p with the 3' splice site during the second step of nuclear pre-mRNA splicing
- PMID: 10684925
- PMCID: PMC111051
- DOI: 10.1093/nar/28.6.1313
Interaction of the yeast DExH-box RNA helicase prp22p with the 3' splice site during the second step of nuclear pre-mRNA splicing
Abstract
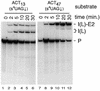
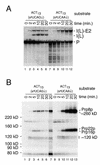
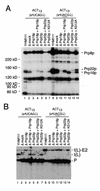
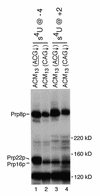
Using site-specific incorporation of the photo-chemical cross-linking reagent 4-thiouridine, we demonstrate the previously unknown association of two proteins with yeast 3' splice sites. One of these is an unidentified approximately 122 kDa protein that cross-links to 3' splice sites during formation of the pre--spliceosome. The other factor is the DExH-box RNA helicase, Prp22p. With substrates functional in the second step of splicing, only very weak cross-linking of Prp22p to intron sequences at the 3' splice site is observed. In contrast, substrates blocked at the second step exhibit strong cross-linking of Prp22 to intron sequences at the 3' splice site, but not to adjacent exon sequences. In vitro reconstitution experiments also show that the association of Prp22p with intron sequences at the 3' splice site is dependent on Prp16p and does not persist when release of mature mRNA from the spliceosome is blocked. Taken together, these results suggest that the 3' splice site of yeast introns is contacted much earlier than previously envisioned by a protein of approximately 120 kDa, and that a transient association of Prp22p with the 3' splice site occurs between the first and second catalytic steps.
Figures

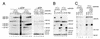
References
-
- Staley J.P. and Guthrie,C. (1998) Cell, 92, 315–326. - PubMed
-
- Moore M.J., Query,C.C. and Sharp,P.A. (1993) In Gesteland,R. and Atkins,J. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–358.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

