Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2'-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs
- PMID: 10684929
- PMCID: PMC111033
- DOI: 10.1093/nar/28.6.1348
Characterisation of the U83 and U84 small nucleolar RNAs: two novel 2'-O-ribose methylation guide RNAs that lack complementarities to ribosomal RNAs
Abstract
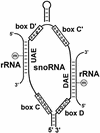
In eukaryotic cells, the site-specific 2'- O -ribose methylation of ribosomal RNAs (rRNAs) and the U6 spliceosomal small nuclear RNA (snRNA) is directed by small nucleolar RNAs (snoRNAs). The C and D box-containing 2'- O -methylation guide snoRNAs select the correct substrate nucleotide through formation of a long 10-21 bp interaction with the target rRNA and U6 snRNA sequences. Here, we report on the characterisation of two novel mammalian C/D box snoRNAs, called U83 and U84, that contain all the elements that are essential for accumulation and function of 2'- O -methylation guide snoRNAs. However, in contrast to all of the known 2'- O -methylation guide RNAs, the human, mouse and pig U83 and U84 snoRNAs feature no antisense elements complementary to rRNA or U6 snRNA sequences. The human U83 and U84 snoRNAs are not associated with maturing nucleolar pre-ribosomal particles, suggesting that they do not function in rRNA biogenesis. Since artificial substrate RNAs complementary to the evolutionarily conserved putative substrate recognition motifs of the U83 and U84 snoRNAs were correctly 2'- O -methylated in the nucleolus of mouse cells, we suggest that the new snoRNAs act as 2'- O -methylation guides for cellular RNAs other then rRNAs and the U6 snRNA.
Figures


References
-
- Hadjiolov A.A. (1985) The Nucleolus and Ribosome Biogenesis. Springer-Verlag, Vienna, Austria.
-
- Maxwell E.S. and Fournier,M.J. (1995) Annu. Rev. Biochem., 64, 897–934. - PubMed
-
- Sollner-Webb B., Tycowski,K.T. and Steitz,J.A. (1996) In Zimmermann,R.A. and Dahlberg,A.E. (eds), Structure, Evolution, Processing and Function in Protein Biosynthesis. CRC Press, Boca Raton, FL, pp. 469–490.
-
- Tollervey D. and Kiss,T. (1997) Curr. Opin. Cell Biol., 9, 337–342. - PubMed
-
- Weinstein L.B. and Steitz,J.A. (1999) Curr. Opin. Cell Biol., 11, 378–384. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

