Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation
- PMID: 10688667
- PMCID: PMC110837
- DOI: 10.1128/MCB.20.6.2209-2217.2000
Fourteen residues of the U1 snRNP-specific U1A protein are required for homodimerization, cooperative RNA binding, and inhibition of polyadenylation
Abstract
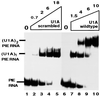
It was previously shown that the human U1A protein, one of three U1 small nuclear ribonucleoprotein-specific proteins, autoregulates its own production by binding to and inhibiting the polyadenylation of its own pre-mRNA. The U1A autoregulatory complex requires two molecules of U1A protein to cooperatively bind a 50-nucleotide polyadenylation-inhibitory element (PIE) RNA located in the U1A 3' untranslated region. Based on both biochemical and nuclear magnetic resonance structural data, it was predicted that protein-protein interactions between the N-terminal regions (amino acids [aa] 1 to 115) of the two U1A proteins would form the basis for cooperative binding to PIE RNA and for inhibition of polyadenylation. In this study, we not only experimentally confirmed these predictions but discovered some unexpected features of how the U1A autoregulatory complex functions. We found that the U1A protein homodimerizes in the yeast two-hybrid system even when its ability to bind RNA is incapacitated. U1A dimerization requires two separate regions, both located in the N-terminal 115 residues. Using both coselection and gel mobility shift assays, U1A dimerization was also observed in vitro and found to depend on the same two regions that were found in vivo. Mutation of the second homodimerization region (aa 103 to 115) also resulted in loss of inhibition of polyadenylation and loss of cooperative binding of two U1A protein molecules to PIE RNA. This same mutation had no effect on the binding of one U1A protein molecule to PIE RNA. A peptide containing two copies of aa 103 to 115 is a potent inhibitor of polyadenylation. Based on these data, a model of the U1A autoregulatory complex is presented.
Figures






References
-
- Allain F H-T, Gubser C C, Howe P W A, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature. 1996;380:646–650. - PubMed
-
- Avis J M, Allain F H-T, Howe P, Varani G, Nagai K, Neuhaus D. Solution structure of the N-terminal RNP domain of U1A protein: the role of the C-terminal residues in structure stability and RNA binding. J Mol Biol. 1996;257:398–411. - PubMed
-
- Boelens W C, Jansen E J R, van Venrooij W J, Stripecke R, Mattaj I W, Gunderson S I. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72:881–892. - PubMed
-
- Boelens W C, Croes Y, de Ruwe M, de Reu L, De Jong W W. Negative charges in the C-terminal domain stabilize the alpha-βcrystallin complex. J Biol Chem. 1998;273:28085–28090. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
