A novel spore peptidoglycan hydrolase of Bacillus cereus: biochemical characterization and nucleotide sequence of the corresponding gene, sleL
- PMID: 10692353
- PMCID: PMC94445
- DOI: 10.1128/JB.182.6.1499-1506.2000
A novel spore peptidoglycan hydrolase of Bacillus cereus: biochemical characterization and nucleotide sequence of the corresponding gene, sleL
Abstract
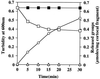
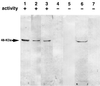
The exudate of germinated spores of B. cereus IFO 13597 in 0.15 M KCl-50 mM potassium phosphate (pH 7.0) contained a spore-lytic enzyme which has substrate specificity for fragmented spore cortex from wild-type organisms (cortical-fragment-lytic enzyme [CFLE]), in addition to a previously characterized germination-specific hydrolase which acts on intact spore cortex (spore cortex-lytic enzyme [SCLE]) (R. Moriyama, S. Kudoh, S. Miyata, S. Nonobe, A. Hattori, and S. Makino, J. Bacteriol. 178:5330-5332, 1996). CFLE was not capable of degrading isolated cortical fragments from spores of Bacillus subtilis ADD1, which lacks muramic acid delta-lactam. This suggests that CFLE cooperates with SCLE in cortex hydrolysis during germination. CFLE was purified in an active form and identified as a 48-kDa protein which functions as an N-acetylglucosaminidase. Immunochemical studies suggested that the mature enzyme is localized on a rather peripheral region of the dormant spore, probably the exterior of the cortex layer. A gene encoding the enzyme, sleL, was cloned in Escherichia coli, and the nucleotide sequence was determined. The gene encodes a protein of 430 amino acids with a deduced molecular weight of 48,136. The N-terminal region contains a repeated motif common to several peptidoglycan binding proteins. Inspection of the data banks showed no similarity of CFLE with N-acetylglucosaminidases found so far, suggesting that CFLE is a novel type of N-acetylglucosaminidase. The B. subtilis genome sequence contains genes, yaaH and ydhD, which encode putative proteins showing similarity to SleL.
Figures




References
-
- Amutha B, Khire J M, Khan M S. Active site characterization of the exo-N-acetyl-β-d-glucosaminidase from thermotolerant Bacillus sp. NCIM 5120: involvement of tryptophan, histidine and carboxylate residues in catalytic activity. Biochim Biophys Acta. 1999;1427:121–132. - PubMed
-
- Atrih A, Bacher G, Körner R, Allmaier G, Foster S J. Structural analysis of Bacillus megaterium KM spore peptidoglycan and its dynamics during germination. Microbiology. 1999;145:1033–1041. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

