Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of recA
- PMID: 10692372
- PMCID: PMC94464
- DOI: 10.1128/JB.182.6.1659-1670.2000
Analysis of Escherichia coli RecA interactions with LexA, lambda CI, and UmuD by site-directed mutagenesis of recA
Abstract
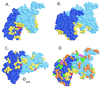
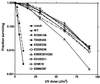
An early event in the induction of the SOS system of Escherichia coli is RecA-mediated cleavage of the LexA repressor. RecA acts indirectly as a coprotease to stimulate repressor self-cleavage, presumably by forming a complex with LexA. How complex formation leads to cleavage is not known. As an approach to this question, it would be desirable to identify the protein-protein interaction sites on each protein. It was previously proposed that LexA and other cleavable substrates, such as phage lambda CI repressor and E. coli UmuD, bind to a cleft located between two RecA monomers in the crystal structure. To test this model, and to map the interface between RecA and its substrates, we carried out alanine-scanning mutagenesis of RecA. Twenty double mutations were made, and cells carrying them were characterized for RecA-dependent repair functions and for coprotease activity towards LexA, lambda CI, and UmuD. One mutation in the cleft region had partial defects in cleavage of CI and (as expected from previous data) of UmuD. Two mutations in the cleft region conferred constitutive cleavage towards CI but not towards LexA or UmuD. By contrast, no mutations in the cleft region or elsewhere in RecA were found to specifically impair the cleavage of LexA. Our data are consistent with binding of CI and UmuD to the cleft between two RecA monomers but do not provide support for the model in which LexA binds in this cleft.
Figures


References
-
- Backman K, Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978;13:65–71. - PubMed
-
- Bailone A, Levine A, Devoret R. Inactivation of prophage λ repressor in vivo. J Mol Biol. 1979;131:553–572. - PubMed
-
- Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. - PubMed
-
- Churchward G, Belin D, Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984;31:165–171. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

