Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages
- PMID: 10698748
- PMCID: PMC91919
- DOI: 10.1128/AEM.66.3.895-903.2000
Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages
Abstract
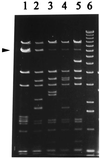
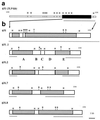
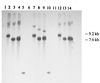
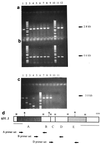
Recombinant phages are generated when Lactococcus lactis subsp. lactis harboring plasmids encoding the abortive type (Abi) of phage resistance mechanisms is infected with small isometric phages belonging to the P335 species. These phage variants are likely to be an important source of virulent new phages that appear in dairy fermentations. They are distinguished from their progenitors by resistance to Abi defenses and by altered genome organization, including regions of L. lactis chromosomal DNA. The objective of this study was to characterize four recombinant variants that arose from infection of L. lactis NCK203 (Abi(+)) with phage phi31. HindIII restriction maps of the variants (phi31.1, phi31.2, phi31.7, and phi31.8) were generated, and these maps revealed the regions containing recombinant DNA. The recombinant region of phage phi31.1, the variant that occurred most frequently, was sequenced and revealed 7.8 kb of new DNA compared with the parent phage, phi31. This region contained numerous instances of homology with various lactococcal temperate phages, as well as homologues of the lambda recombination protein BET and Escherichia coli Holliday junction resolvase Rus, factors which may contribute to efficient recombination processes. A sequence analysis and phenotypic tests revealed a new origin of replication in the phi31.1 DNA, which replaced the phi31 origin. Three separate HindIII fragments, accounting for most of the recombinant region of phi31.1, were separately cloned into gram-positive suicide vector pTRK333 and transformed into NCK203. Chromosomal insertions of each plasmid prevented the appearance of different combinations of recombinant phages. The chromosomal insertions did not affect an inducible prophage present in NCK203. Our results demonstrated that recombinant phages can acquire DNA cassettes from different regions of the chromosome in order to overcome Abi defenses. Disruption of these regions by insertion can alter the types and diversity of new phages that appear during phage-host interactions.
Figures


References
-
- Birkeland N-K, Lonneborg A-M. The cos region of lactococcal bacteriophage φLC3. DNA Sequence J. 1993;4:211–214. - PubMed
-
- Chandry P S, Moore S C, Boyce J D, Davidson B E, Hillier A J. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol. 1997;26:49–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

