Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes
- PMID: 10698754
- PMCID: PMC91925
- DOI: 10.1128/AEM.66.3.930-936.2000
Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes
Abstract
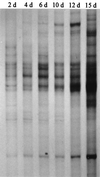
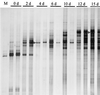
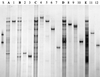
A cultivation-independent technique for genetic profiling of PCR-amplified small-subunit rRNA genes (SSU rDNA) was chosen to characterize the diversity and succession of microbial communities during composting of an organic agricultural substrate. PCR amplifications were performed with DNA directly extracted from compost samples and with primers targeting either (i) the V4-V5 region of eubacterial 16S rRNA genes, (ii) the V3 region in the 16S rRNA genes of actinomycetes, or (iii) the V8-V9 region of fungal 18S rRNA genes. Homologous PCR products were converted to single-stranded DNA molecules by exonuclease digestion and were subsequently electrophoretically separated by their single-strand-conformation polymorphism (SSCP). Genetic profiles obtained by this technique showed a succession and increasing diversity of microbial populations with all primers. A total of 19 single products were isolated from the profiles by PCR reamplification and cloning. DNA sequencing of these molecular isolates showed similarities in the range of 92.3 to 100% to known gram-positive bacteria with a low or high G+C DNA content and to the SSU rDNA of gamma-Proteobacteria. The amplified 18S rRNA gene sequences were related to the respective gene regions of Candida krusei and Candida tropicalis. Specific molecular isolates could be attributed to different composting stages. The diversity of cultivated bacteria isolated from samples taken at the end of the composting process was low. A total of 290 isolates were related to only 6 different species. Two or three of these species were also detectable in the SSCP community profiles. Our study indicates that community SSCP profiles can be highly useful for the monitoring of bacterial diversity and community successions in a biotechnologically relevant process.
Figures

References
-
- Andrews S A, Lee H, Trevors J T. Bacterial species in raw and cured compost from a large-scale urban composter. J Ind Microbiol. 1994;13:177–182.
-
- Bassam B J, Caetano-Anolles G, Greshoff P M. Fast and sensitive staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. - PubMed
-
- Beffa T, Blanc M, Aragno M. Obligately and facultatively autotrophic, sulfur- and hydrogen-oxidizing thermophilic bacteria isolated from hot composts. Arch Microbiol. 1996;165:34–40.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

