Genetic and biochemical diversity among isolates of Paenibacillus alvei cultured from Australian honeybee (Apis mellifera) colonies
- PMID: 10698777
- PMCID: PMC91948
- DOI: 10.1128/AEM.66.3.1098-1106.2000
Genetic and biochemical diversity among isolates of Paenibacillus alvei cultured from Australian honeybee (Apis mellifera) colonies
Abstract
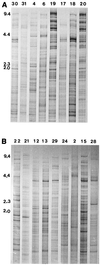
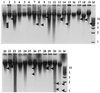
Twenty-five unique CfoI-generated whole-cell DNA profiles were identified in a study of 30 Paenibacillus alvei isolates cultured from honey and diseased larvae collected from honeybee (Apis mellifera) colonies in geographically diverse areas in Australia. The fingerprint patterns were highly variable and readily discernible from one another, which highlighted the potential of this method for tracing the movement of isolates in epidemiological studies. 16S rRNA gene fragments (length, 1,416 bp) for all 30 isolates were enzymatically amplified by PCR and subjected to restriction analysis with DraI, HinfI, CfoI, AluI, FokI, and RsaI. With each enzyme the restriction profiles of the 16S rRNA genes from all 30 isolates were identical (one restriction fragment length polymorphism [RFLP] was observed in the HinfI profile of the 16S rRNA gene from isolate 17), which confirmed that the isolates belonged to the same species. The restriction profiles generated by using DraI, FokI, and HinfI differentiated P. alvei from the phylogenetically closely related species Paenibacillus macerans and Paenibacillus macquariensis. Alveolysin gene fragments (length, 1, 555 bp) were enzymatically amplified from some of the P. alvei isolates (19 of 30 isolates), and RFLP were detected by using the enzymes CfoI, Sau3AI, and RsaI. Extrachromosomal DNA ranging in size from 1 to 10 kb was detected in 17 of 30 (57%) P. alvei whole-cell DNA profiles. Extensive biochemical heterogeneity was observed among the 28 P. alvei isolates examined with the API 50CHB system. All of these isolates were catalase, oxidase, and Voges-Proskauer positive and nitrate negative, and all produced acid when glycerol, esculin, and maltose were added. The isolates produced variable results for 16 of the 49 biochemical tests; negative reactions were recorded in the remaining 30 assays. The genetic and biochemical heterogeneity in P. alvei isolates may be a reflection of adaptation to the special habitats in which they originated.
Figures





Similar articles
-
Geographically diverse Australian isolates of Melissococcus pluton exhibit minimal genotypic diversity by restriction endonuclease analysis.FEMS Microbiol Lett. 1999 Apr 15;173(2):311-8. doi: 10.1111/j.1574-6968.1999.tb13519.x. FEMS Microbiol Lett. 1999. PMID: 10227161
-
Feasibility of using RFLP of PCR-amplified 16S rRNA gene(s) for rapid differentiation of isolates of aerobic spore-forming bacteria from honey.J Microbiol Methods. 2019 Oct;165:105690. doi: 10.1016/j.mimet.2019.105690. Epub 2019 Aug 16. J Microbiol Methods. 2019. PMID: 31425714
-
Distinct differentiation of closely related species of Bacillus subtilis group with industrial importance.J Microbiol Methods. 2011 Nov;87(2):161-4. doi: 10.1016/j.mimet.2011.08.011. Epub 2011 Aug 26. J Microbiol Methods. 2011. PMID: 21889958
-
Differentiation of Paenibacillus larvae subsp. larvae, the cause of American foulbrood of honeybees, by using PCR and restriction fragment analysis of genes encoding 16S rRNA.Appl Environ Microbiol. 2002 Jul;68(7):3655-60. doi: 10.1128/AEM.68.7.3655-3660.2002. Appl Environ Microbiol. 2002. PMID: 12089057 Free PMC article.
-
Genetic and biochemical diversity of Paenibacillus larvae isolated from Tunisian infected honey bee broods.Biomed Res Int. 2013;2013:479893. doi: 10.1155/2013/479893. Epub 2013 Sep 2. Biomed Res Int. 2013. PMID: 24073406 Free PMC article.
Cited by
-
Isolation and identification of a new intracellular antimicrobial peptide produced by Paenibacillus alvei AN5.World J Microbiol Biotechnol. 2014 Apr;30(4):1377-85. doi: 10.1007/s11274-013-1558-z. Epub 2013 Nov 24. World J Microbiol Biotechnol. 2014. PMID: 24272828
-
A novel bacterial pathogen of Biomphalaria glabrata: a potential weapon for schistosomiasis control?PLoS Negl Trop Dis. 2015 Feb 26;9(2):e0003489. doi: 10.1371/journal.pntd.0003489. eCollection 2015 Feb. PLoS Negl Trop Dis. 2015. PMID: 25719489 Free PMC article.
-
Extracts of Talaromyces purpureogenus Strains from Apis mellifera Bee Bread Inhibit the Growth of Paenibacillus spp. In Vitro.Microorganisms. 2023 Aug 11;11(8):2067. doi: 10.3390/microorganisms11082067. Microorganisms. 2023. PMID: 37630627 Free PMC article.
-
Phylogeny in aid of the present and novel microbial lineages: diversity in Bacillus.PLoS One. 2009;4(2):e4438. doi: 10.1371/journal.pone.0004438. Epub 2009 Feb 12. PLoS One. 2009. PMID: 19212464 Free PMC article.
-
Characterization of toxin systems of Paenibacillus strains isolated from honeybees.Sci Rep. 2025 Aug 26;15(1):31346. doi: 10.1038/s41598-025-12956-x. Sci Rep. 2025. PMID: 40858718 Free PMC article.
References
-
- Ash C, Farrow J A E, Wallbanks S, Collins M D. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett Appl Microbiol. 1991;13:202–206.
-
- Ash C, Priest F G, Collins M D. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Antonie Leewenhoek J Microbiol. 1993;64:253–260. - PubMed
-
- Bailey L, Ball B V. Honey bee pathology. 2nd ed. Sidcup, United Kingdom: Harcourt Brace Jovanovich; 1991. pp. 36–41.
-
- Balaraman K, Rao U S B, Rajagopalan P K. Bacterial pathogens of mosquito larvae—Bacillus alvei (Cheshire and Cheyene) and Bacillus brevis (Migula) isolated in Pondicherry. Indian J Med Res. 1979;70:615–619. - PubMed
-
- Cowan S T, Steele K J. Appendix C: characterisation tests. In: Barrow G I, Feltham R K A, editors. Manual for the identification of medical bacteria. Cambridge, United Kingdom: Cambridge University Press; 1993. pp. 219–238.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

