Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation
- PMID: 10698788
- PMCID: PMC91959
- DOI: 10.1128/AEM.66.3.1175-1182.2000
Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation
Abstract
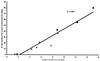
Laboratory-scale sequencing batch reactors (SBRs) as models for activated sludge processes were used to study enhanced biological phosphorus removal (EBPR) from wastewater. Enrichment for polyphosphate-accumulating organisms (PAOs) was achieved essentially by increasing the phosphorus concentration in the influent to the SBRs. Fluorescence in situ hybridization (FISH) using domain-, division-, and subdivision-level probes was used to assess the proportions of microorganisms in the sludges. The A sludge, a high-performance P-removing sludge containing 15.1% P in the biomass, was comprised of large clusters of polyphosphate-containing coccobacilli. By FISH, >80% of the A sludge bacteria were beta-2 Proteobacteria arranged in clusters of coccobacilli, strongly suggesting that this group contains a PAO responsible for EBPR. The second dominant group in the A sludge was the Actinobacteria. Clone libraries of PCR-amplified bacterial 16S rRNA genes from three high-performance P-removing sludges were prepared, and clones belonging to the beta-2 Proteobacteria were fully sequenced. A distinctive group of clones (sharing >/=98% sequence identity) related to Rhodocyclus spp. (94 to 97% identity) and Propionibacter pelophilus (95 to 96% identity) was identified as the most likely candidate PAOs. Three probes specific for the highly related candidate PAO group were designed from the sequence data. All three probes specifically bound to the morphologically distinctive clusters of PAOs in the A sludge, exactly coinciding with the beta-2 Proteobacteria probe. Sequential FISH and polyphosphate staining of EBPR sludges clearly demonstrated that PAO probe-binding cells contained polyphosphate. Subsequent PAO probe analyses of a number of sludges with various P removal capacities indicated a strong positive correlation between P removal from the wastewater as determined by sludge P content and number of PAO probe-binding cells. We conclude therefore that an important group of PAOs in EBPR sludges are bacteria closely related to Rhodocyclus and Propionibacter.
Figures



References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Blackall L L. Molecular identification of activated sludge foaming bacteria. Water Sci Technol. 1994;29(7):35–42.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

