Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis
- PMID: 10698924
- PMCID: PMC305622
- DOI: 10.1093/emboj/19.5.819
Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis
Abstract
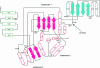
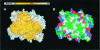
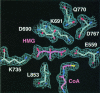
3-hydroxy-3-methylglutaryl-CoA reductase (HMGR) catalyzes the formation of mevalonate, the committed step in the biosynthesis of sterols and isoprenoids. The activity of HMGR is controlled through synthesis, degradation and phosphorylation to maintain the concentration of mevalonate-derived products. In addition to the physiological regulation of HMGR, the human enzyme has been targeted successfully by drugs in the clinical treatment of high serum cholesterol levels. Three crystal structures of the catalytic portion of human HMGR in complexes with HMG-CoA, with HMG and CoA, and with HMG, CoA and NADP(+), provide a detailed view of the enzyme active site. Catalytic portions of human HMGR form tight tetramers. The crystal structure explains the influence of the enzyme's oligomeric state on the activity and suggests a mechanism for cholesterol sensing. The active site architecture of human HMGR is different from that of bacterial HMGR; this may explain why binding of HMGR inhibitors to bacterial HMGRs has not been reported.
Figures


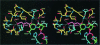
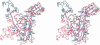
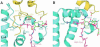
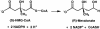
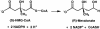
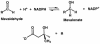
References
-
- Bennis F., Favre, G., Le Gaillard, F. and Soula, G. (1993) Importance of mevalonate-derived products in the control of HMG-CoA reductase activity and growth of human lung adenocarcinoma cell line A549. Int. J. Cancer, 55, 640–645. - PubMed
-
- Blackwell J.R. and Horgan, R. (1991) A novel strategy for production of a highly expressed recombinant protein in an active form. FEBS Lett., 295, 10–12. - PubMed
-
- Bochar D.A., Stauffacher, C.V. and Rodwell, V.W. (1999a) Sequence comparisons reveal two classes of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Genet. Metab., 66, 122–127. - PubMed
-
- Bochar D.A., Tabernero, L., Stauffacher, C.V. and Rodwell, V.W. (1999b) Aminoethylcysteine can replace the function of the essential active site lysine of Pseudomonas mevalonii 3-hydroxy-3-methylglutaryl coenzyme A reductase. Biochemistry, 38, 8879–8883. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

