Tumorigenesis in mice with a fusion of the leukaemia oncogene Mll and the bacterial lacZ gene
- PMID: 10698926
- PMCID: PMC305624
- DOI: 10.1093/emboj/19.5.843
Tumorigenesis in mice with a fusion of the leukaemia oncogene Mll and the bacterial lacZ gene
Abstract
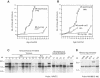
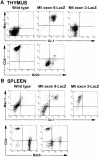
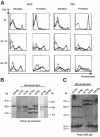
Many different chromosomal translocations occur in man at chromosome 11q23 in acute leukaemias. Molecular analyses revealed that the MLL gene (also called ALL-1, HRX or HTRX) is broken by the translocations, causing fusion with genes from other chromosomes. The diversity of MLL fusion partners poses a dilemma about the function of the fusion proteins in tumour development. The consequence of MLL truncation and fusion has been analysed by joining exon 8 of Mll with the bacterial lacZ gene using homologous recombination in mouse embryonic stem cells. We show that this fusion is sufficient to cause embryonic stem cell-derived acute leukaemias in chimeric mice, and these tumours occur with long latency compared with those found in MLL-Af9 chimeric mice. These findings indicate that an MLL fusion protein can contribute to tumorigenesis, even if the fusion partner has no known pathogenic role. Thus, truncation and fusion of MLL can be sufficient for tumorigenesis, regardless of the fusion partner.
Figures



References
-
- Atlas M., Head, D., Behm, F., Schmidt, E., Zeleznik-Le, N.J., Roe, B.A., Burian, D. and Domer, P.H. (1998) Cloning and sequence analysis of four t(9;11) therapy-related leukemia breakpoints. Leukemia, 12, 1895–1902. - PubMed
-
- Bernard O. and Berger, R. (1995) Molecular basis of 11q23 rearrangements in hematopoietic malignant proliferations. Genes Chromosomes Cancer, 13, 75–85. - PubMed
-
- Chaplin T., et al. (1995a) A novel class of zinc finger/leucine zipper genes identified from the molecular cloning of the t(10;11) translocation in acute leukemia. Blood, 85, 1435–1441. - PubMed
-
- Chaplin T., Bernard, O., Beverloo, H.B., Saha, V., Hagemeijer, A., Berger, R. and Young, B.D. (1995b) The t(10;11) translocation in acute myeloid leukemia (M5) consistently fuses the leucine zipper motif of AF10 onto the HRX gene. Blood, 86, 2073–2076. - PubMed
-
- Cimino C., et al. (1997) Infant acute leukemias show the same biased distribution of ALL1 gene breaks as topoisomerase II related secondary acute leukemias. Cancer Res., 57, 2879–2883. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

