Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability
- PMID: 10698950
- PMCID: PMC305648
- DOI: 10.1093/emboj/19.5.1098
Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability
Abstract
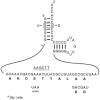
tmRNA, through its tRNA and mRNA properties, adds short peptide tags to abnormal proteins, targeting these proteins for proteolytic degradation. Although the conservation of tmRNA throughout the bacterial kingdom suggests that it must provide a strong selective advantage, it has not been shown to be essential for any bacterium. We report that tmRNA is essential in Neisseria gonorrhoeae. Although tagging per se appears to be required for gonococcal viability, tagging for proteolysis does not. This suggests that the essential roles of tmRNA in N.gonorrhoeae may include resolving stalled translation complexes and/or preventing depletion of free ribosomes. Although derivatives of N.gonorrhoeae expressing Escherichia coli tmRNA as their sole tmRNA were isolated, they appear to form colonies only after acquiring an extragenic suppressor(s).
Figures



References
-
- Botstein D. and Herskowits, I. (1974) Properties of hybrids between Salmonella phage P22 and coliphage lambda. Nature, 251, 584–589. - PubMed
-
- Bott K., Stewart,G.C. and Anderson,A.G. (1984) Genetic mapping of cloned ribosomal RNA genes. In Hoch,J.A. and Ganesan,A.T. (eds), Syntro Conference on Genetics and Biotechnology of Bacilli. Academic Press, New York, NY, pp. 19–34.
-
- Britigan B.E., Cohen, M.S. and Sparling, P.F. (1985) Gonococcal infection: a model of molecular pathogenesis. N. Engl. J. Med., 312, 1683–1694. - PubMed
-
- Chauhan A.K. and Apirion, D. (1989) The gene for a small stable RNA (10Sa RNA) of Escherichia coli. Mol. Microbiol., 3, 1481–1485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

