Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications
- PMID: 10698952
- PMCID: PMC305650
- DOI: 10.1093/emboj/19.5.1119
Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications
Abstract
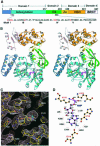
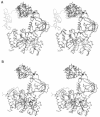
DNA ligases catalyze the crucial step of joining the breaks in duplex DNA during DNA replication, repair and recombination, utilizing either ATP or NAD(+) as a cofactor. Despite the difference in cofactor specificity and limited overall sequence similarity, the two classes of DNA ligase share basically the same catalytic mechanism. In this study, the crystal structure of an NAD(+)-dependent DNA ligase from Thermus filiformis, a 667 residue multidomain protein, has been determined by the multiwavelength anomalous diffraction (MAD) method. It reveals highly modular architecture and a unique circular arrangement of its four distinct domains. It also provides clues for protein flexibility and DNA-binding sites. A model for the multidomain ligase action involving large conformational changes is proposed.
Figures
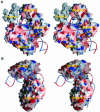

Similar articles
-
DNA ligases in the repair and replication of DNA.Mutat Res. 2000 Aug 30;460(3-4):301-18. doi: 10.1016/s0921-8777(00)00033-1. Mutat Res. 2000. PMID: 10946235 Review.
-
Structure of the adenylation domain of an NAD+-dependent DNA ligase.Structure. 1999 Jan 15;7(1):35-42. doi: 10.1016/s0969-2126(99)80007-0. Structure. 1999. PMID: 10368271
-
Identification of essential residues in Thermus thermophilus DNA ligase.Nucleic Acids Res. 1996 Aug 1;24(15):3079-85. doi: 10.1093/nar/24.15.3079. Nucleic Acids Res. 1996. PMID: 8760897 Free PMC article.
-
Novel DNA ligase with broad nucleotide cofactor specificity from the hyperthermophilic crenarchaeon Sulfophobococcus zilligii: influence of ancestral DNA ligase on cofactor utilization.Environ Microbiol. 2008 Dec;10(12):3212-24. doi: 10.1111/j.1462-2920.2008.01710.x. Epub 2008 Jul 16. Environ Microbiol. 2008. PMID: 18647334
-
Structural and mechanistic conservation in DNA ligases.Nucleic Acids Res. 2000 Nov 1;28(21):4051-8. doi: 10.1093/nar/28.21.4051. Nucleic Acids Res. 2000. PMID: 11058099 Free PMC article. Review.
Cited by
-
Biochemical and Structural Characterisation of DNA Ligases from Bacteria and Archaea.Biosci Rep. 2016 Oct 6;36(5):00391. doi: 10.1042/BSR20160003. Biosci Rep. 2016. PMID: 27582505 Free PMC article.
-
The NAD-dependent ligase encoded by yerG is an essential gene of Bacillus subtilis.Nucleic Acids Res. 2000 Dec 1;28(23):4642-8. doi: 10.1093/nar/28.23.4642. Nucleic Acids Res. 2000. PMID: 11095673 Free PMC article.
-
A second NAD(+)-dependent DNA ligase (LigB) in Escherichia coli.Nucleic Acids Res. 2001 Dec 15;29(24):4930-4. doi: 10.1093/nar/29.24.4930. Nucleic Acids Res. 2001. PMID: 11812821 Free PMC article.
-
Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD+-dependent polynucleotide ligases.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2592-2597. doi: 10.1073/pnas.1619220114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223499 Free PMC article.
-
Characterisation and engineering of a thermophilic RNA ligase from Palaeococcus pacificus.Nucleic Acids Res. 2024 Apr 24;52(7):3924-3937. doi: 10.1093/nar/gkae149. Nucleic Acids Res. 2024. PMID: 38421610 Free PMC article.
References
-
- Aravind L. and Koonin, E.V. (1999) Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol., 287, 1023–1040. - PubMed
-
- Bochkarev A., Pfuetzner, R.A., Edwards, A.M. and Frappier, L. (1997) Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature, 385, 176–181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

