Cryptococcus neoformans STE12alpha regulates virulence but is not essential for mating
- PMID: 10704467
- PMCID: PMC2195848
- DOI: 10.1084/jem.191.5.871
Cryptococcus neoformans STE12alpha regulates virulence but is not essential for mating
Abstract
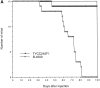
The Cryptococcus neoformans STE12alpha gene, a homologue of Saccharomyces cerevisiae STE12, exists only in mating type (MAT)alpha cells. In S. cerevisiae, STE12 was required for mating and filament formation. In C. neoformans, haploid fruiting on filament agar required STE12alpha. The ability to form hyphae, however, was not affected by deletion of STE12alpha when convergently growing MATa strains were present. Furthermore, ste12alpha disruptants were fertile when mated with MATa strains, albeit with reduced mating frequency. Most importantly, the virulence of a ste12alpha disruptant of serotype D strain was significantly reduced in a mouse model. When the ste12alpha locus was reconstituted with the wild-type allele by cotransformation, virulence was restored. Histopathological analysis demonstrated a reduction in capsular size of yeast cells, less severe cystic lesions, and stronger immune responses in meninges of mice infected with ste12alpha cells than those of mice infected with STE12alpha cells. Using reporter gene constructs, we found that STE12alpha controls the expression of several phenotypes known to be involved in virulence, such as capsule and melanin production. These results demonstrate a clear molecular link between mating type and virulence in C. neoformans.
Figures









Similar articles
-
The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans.Genetics. 1999 Dec;153(4):1601-15. doi: 10.1093/genetics/153.4.1601. Genetics. 1999. PMID: 10581270 Free PMC article.
-
The second STE12 homologue of Cryptococcus neoformans is MATa-specific and plays an important role in virulence.Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):3258-63. doi: 10.1073/pnas.061031998. Epub 2001 Feb 27. Proc Natl Acad Sci U S A. 2001. PMID: 11248066 Free PMC article.
-
The Cryptococcus neoformans STE12alpha gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific.Mol Microbiol. 1997 Dec;26(5):951-60. doi: 10.1046/j.1365-2958.1997.6322001.x. Mol Microbiol. 1997. PMID: 9426132
-
[Mating types, sexual reproduction and ploidy in fungi: effects on virulence].Mikrobiyol Bul. 2009 Jul;43(3):507-13. Mikrobiyol Bul. 2009. PMID: 19795629 Review. Turkish.
-
Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans.Fungal Genet Biol. 1998 Oct;25(1):1-14. doi: 10.1006/fgbi.1998.1079. Fungal Genet Biol. 1998. PMID: 9806801 Review.
Cited by
-
Cryptococcus neoformans {alpha} strains preferentially disseminate to the central nervous system during coinfection.Infect Immun. 2005 Aug;73(8):4922-33. doi: 10.1128/IAI.73.8.4922-4933.2005. Infect Immun. 2005. PMID: 16041006 Free PMC article.
-
Identification of QTLs Associated with Virulence Related Traits and Drug Resistance in Cryptococcus neoformans.G3 (Bethesda). 2016 Sep 8;6(9):2745-59. doi: 10.1534/g3.116.029595. G3 (Bethesda). 2016. PMID: 27371951 Free PMC article.
-
Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans.Eukaryot Cell. 2004 Feb;3(1):14-26. doi: 10.1128/EC.3.1.14-26.2004. Eukaryot Cell. 2004. PMID: 14871933 Free PMC article.
-
Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14455-60. doi: 10.1073/pnas.97.26.14455. Proc Natl Acad Sci U S A. 2000. PMID: 11121047 Free PMC article.
-
A Wor1-Like Transcription Factor Is Essential for Virulence of Cryptococcus neoformans.Front Cell Infect Microbiol. 2018 Nov 13;8:369. doi: 10.3389/fcimb.2018.00369. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30483479 Free PMC article.
References
-
- Kwon-Chung K.J., Bennett J.E. Medical Mycology 1992. Lea & Febiger; Philadelphia: pp. 397–446
-
- Kwon-Chung K.J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans . Mycologia. 1975;67:1197–1200. - PubMed
-
- Kwon-Chung K.J. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–946. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

