Higher levels of organization in the interphase nucleus of cycling and differentiated cells
- PMID: 10704477
- PMCID: PMC98989
- DOI: 10.1128/MMBR.64.1.138-152.2000
Higher levels of organization in the interphase nucleus of cycling and differentiated cells
Abstract
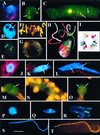
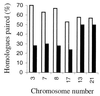
The review examines the structured organization of interphase nuclei using a range of examples from the plants, animals, and fungi. Nuclear organization is shown to be an important phenomenon in cell differentiation and development. The review commences by examining nuclei in dividing cells and shows that the organization patterns can be dynamic within the time frame of the cell cycle. When cells stop dividing, derived differentiated cells often show quite different nuclear organizations. The developmental fate of nuclei is divided into three categories. (i) The first includes nuclei that undergo one of several forms of polyploidy and can themselves change in structure during the course of development. Possible function roles of polyploidy is given. (ii) The second is nuclear reorganization without polyploidy, where nuclei reorganize their structure to form novel arrangements of proteins and chromosomes. (iii) The third is nuclear disintegration linked to programmed cell death. The role of the nucleus in this process is described. The review demonstrates that recent methods to probe nuclei for nucleic acids and proteins, as well as to examine their intranuclear distribution in vivo, has revealed much about nuclear structure. It is clear that nuclear organization can influence or be influenced by cell activity and development. However, the full functional role of many of the observed phenomena has still to be fully realized.
Figures



References
-
- Anamthawat-Jönsson K, Heslop-Harrison J S. Centromeres, telomeres and chromatin in the interphase nucleus of cereals. Caryologia. 1990;43:205–213.
-
- Appels R, Honeycutt R L. rDNA: evolution over a billion years. In: Datta S K, editor. DNA systematics. II. Plants. Boca Raton, Fla: CRC Press Inc.; 1986. pp. 81–135.
-
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromeric sequence. Chromosoma. 1996;105:261–268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

