Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process
- PMID: 10704480
- PMCID: PMC98992
- DOI: 10.1128/MMBR.64.1.202-236.2000
Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process
Abstract
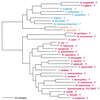
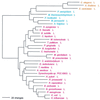
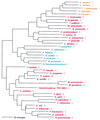
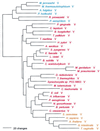
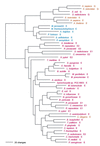
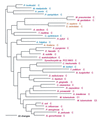
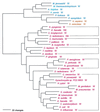
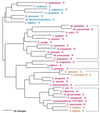
The aminoacyl-tRNA synthetases (AARSs) and their relationship to the genetic code are examined from the evolutionary perspective. Despite a loose correlation between codon assignments and AARS evolutionary relationships, the code is far too highly structured to have been ordered merely through the evolutionary wanderings of these enzymes. Nevertheless, the AARSs are very informative about the evolutionary process. Examination of the phylogenetic trees for each of the AARSs reveals the following. (i) Their evolutionary relationships mostly conform to established organismal phylogeny: a strong distinction exists between bacterial- and archaeal-type AARSs. (ii) Although the evolutionary profiles of the individual AARSs might be expected to be similar in general respects, they are not. It is argued that these differences in profiles reflect the stages in the evolutionary process when the taxonomic distributions of the individual AARSs became fixed, not the nature of the individual enzymes. (iii) Horizontal transfer of AARS genes between Bacteria and Archaea is asymmetric: transfer of archaeal AARSs to the Bacteria is more prevalent than the reverse, which is seen only for the "gemini group. " (iv) The most far-ranging transfers of AARS genes have tended to occur in the distant evolutionary past, before or during formation of the primary organismal domains. These findings are also used to refine the theory that at the evolutionary stage represented by the root of the universal phylogenetic tree, cells were far more primitive than their modern counterparts and thus exchanged genetic material in far less restricted ways, in effect evolving in a communal sense.
Figures


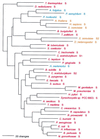
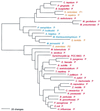
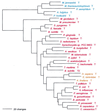
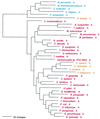
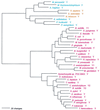
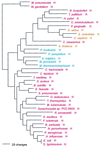


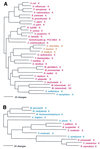
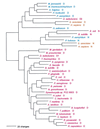
References
-
- Adduce J, Hoosegow M. MOLPHY: programs for maximum likelihood inference of protein phylogeny. Computer science monographs, no. 27. Tokyo, Japan: Institute of Statistical Mathematics; 1992.
-
- Andersson S G, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C, Podowski R M, Naslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
-
- Arnez J G, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci. 1997;22:211–216. - PubMed
-
- Berthet-Colominas C, Seignovert L, Härtlein M, Grotli M, Cusack S, Leberman R. The crystal structure of asparaginyl-tRNA synthetase from Thermus thermophilus and its complexes with ATP and asparaginyl-adenylate: the mechanism of discrimination between asparagine and aspartic acid. EMBO J. 1998;17:2947–2960. - PMC - PubMed
-
- Brown J R. Aminoacyl-tRNA synthetases: evolution of a troubled family. In: Wiegel J, Adams M H W, editors. Thermophiles: the keys to molecular evolution and the origin of life? London, United Kingdom: Taylor & Francis; 1998. pp. 217–230.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

