The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates
- PMID: 10706615
- PMCID: PMC16022
- DOI: 10.1073/pnas.040580197
The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates
Abstract
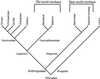
Olfactory receptors (ORs) located in the cell membrane of olfactory sensory neurons of the nasal epithelium are responsible for odor detection by binding specific odorant ligands. Primates are thought to have a reduced sense of smell (microsmatic) with respect to other mammals such as dogs or rodents. We have previously demonstrated that over 70% of the human OR genes have become nonfunctional pseudogenes, leading us to hypothesize that the reduced sense of smell could correlate with the loss of functional genes. To extend these results, we sampled the OR gene repertoire of 10 primate species, from prosimian lemur to human, in addition to mouse. About 221 previously unidentified primate sequences and 33 mouse sequences were analyzed. These sequences encode ORs distributed in seven families and 56 subfamilies. Analysis showed a high fraction ( approximately 50% on average) of pseudogenes in hominoids. In contrast, only approximately 27% of OR genes are pseudogenes in Old World monkeys, and New World monkeys are almost free of pseudogenes. The prosimian branch seems to have evolved differently from the other primates and has approximately 37% pseudogene content. No pseudogenes were found in mouse. With the exception of New World monkeys, we demonstrate that primates have a high fraction of OR pseudogenes compared with mouse. We hypothesize that under relaxed selective constraints, primates would have progressively accumulated pseudogenes with the highest level seen in hominoids. The fraction of pseudogenes in the OR gene repertoire could parallel the evolution of the olfactory sensory function.
Figures

Similar articles
-
Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision.Mol Biol Evol. 2010 May;27(5):1192-200. doi: 10.1093/molbev/msq003. Epub 2010 Jan 8. Mol Biol Evol. 2010. PMID: 20061342
-
The canine olfactory subgenome.Genomics. 2004 Mar;83(3):361-72. doi: 10.1016/j.ygeno.2003.08.009. Genomics. 2004. PMID: 14962662
-
Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes.Genomics. 1999 Oct 1;61(1):24-36. doi: 10.1006/geno.1999.5900. Genomics. 1999. PMID: 10512677
-
Genetic basis of olfactory communication in primates.Am J Primatol. 2006 Jun;68(6):559-67. doi: 10.1002/ajp.20252. Am J Primatol. 2006. PMID: 16715504 Review.
-
Odorant receptor genes in humans.Curr Opin Genet Dev. 1999 Jun;9(3):315-20. doi: 10.1016/s0959-437x(99)80047-1. Curr Opin Genet Dev. 1999. PMID: 10377291 Review.
Cited by
-
A comparison of reptilian and avian olfactory receptor gene repertoires: species-specific expansion of group gamma genes in birds.BMC Genomics. 2009 Sep 21;10:446. doi: 10.1186/1471-2164-10-446. BMC Genomics. 2009. PMID: 19772566 Free PMC article.
-
Strong links between genomic and anatomical diversity in both mammalian olfactory chemosensory systems.Proc Biol Sci. 2014 Apr 9;281(1783):20132828. doi: 10.1098/rspb.2013.2828. Print 2014 May 22. Proc Biol Sci. 2014. PMID: 24718758 Free PMC article.
-
Birds Generally Carry a Small Repertoire of Bitter Taste Receptor Genes.Genome Biol Evol. 2015 Sep 4;7(9):2705-15. doi: 10.1093/gbe/evv180. Genome Biol Evol. 2015. PMID: 26342138 Free PMC article.
-
Stop and Smell the Pollen: The Role of Olfaction and Vision of the Oriental Honey Buzzard in Identifying Food.PLoS One. 2015 Jul 15;10(7):e0130191. doi: 10.1371/journal.pone.0130191. eCollection 2015. PLoS One. 2015. PMID: 26177533 Free PMC article.
-
What does the nose know? Olfactory function predicts social network size in human.Sci Rep. 2016 Apr 25;6:25026. doi: 10.1038/srep25026. Sci Rep. 2016. PMID: 27109506 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

