Detection of leukemia-associated MLL-GAS7 translocation early during chemotherapy with DNA topoisomerase II inhibitors
- PMID: 10706619
- PMCID: PMC16012
- DOI: 10.1073/pnas.050397097
Detection of leukemia-associated MLL-GAS7 translocation early during chemotherapy with DNA topoisomerase II inhibitors
Abstract
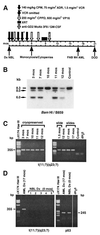
Leukemias with MLL gene translocations are a complication of primary cancer treatment with DNA topoisomerase II inhibitors. How early translocations appear during primary cancer treatment has not been investigated. We tracked the leukemic clone with an MLL gene translocation during neuroblastoma therapy in a child who developed acute myeloid leukemia. The karyotype of the leukemic clone showed del(11)(q23). We used panhandle PCR-based methods to isolate the breakpoint junction involving MLL and an unknown partner gene. Marrow DNA from neuroblastoma diagnosis and DNA and RNA from serial preleukemic marrows were examined for the translocation. The karyotypic del(11)(q23) was a cryptic t(11;17). GAS7, a growth arrest-specific gene at chromosome band 17p13, was the partner gene of MLL. Two different MLL-GAS7 fusion transcripts were expressed. The translocation was already detectable by 1.5 months after the start of neuroblastoma treatment. The translocation was not detectable in the marrow at neuroblastoma diagnosis or in peripheral blood lymphocyte DNAs of six normal subjects. GAS7 is a new partner gene of MLL in treatment-related acute myeloid leukemia. MLL gene translocations can be present early during anticancer treatment at low cumulative doses of DNA topoisomerase II inhibitors. Although MLL has many partner genes and most have not been characterized, panhandle PCR strategies afford new means for detecting MLL gene translocations early during therapy when the partner gene is unknown.
Figures


References
-
- Felix C A. Biochim Biophys Acta. 1998;1400:233–255. - PubMed
-
- Kushner B H, Cheung N K, Kramer K, Heller G, Jhanwar S C. J Clin Oncol. 1998;16:3880–3889. - PubMed
-
- Kushner B H, Heller G, Cheung N-K V, Wollner N, Kramer K, Bajorin D, Polyak T, Meyers P A. J Clin Oncol. 1998;16:3016–3020. - PubMed
-
- Smith M, Rubenstein L, Anderson J, Arthur D, Catalano P, Freidlin B, Heyn R, Khayat A, Krailo M, Land V, et al. J Clin Oncol. 1999;17:569–577. - PubMed
-
- Meadows A T, Obringer A C, Marrero O, Oberlin O, Robison L, Fossati-Bellani F, Green D, Voute P A, Morris-Jones P, Greenburg M, et al. Med Pediatr Oncol. 1989;17:477–484. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

