Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA
- PMID: 10708419
- PMCID: PMC111803
- DOI: 10.1128/jvi.74.7.3046-3057.2000
Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA
Abstract
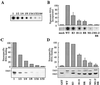
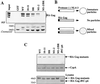
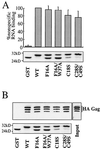
Retroviral Gag polyproteins drive virion assembly by polymerizing to form a spherical shell that lines the inner membrane of nascent virions. Deletion of the nucleocapsid (NC) domain of the Gag polyprotein disrupts assembly, presumably because NC is required for polymerization. Human immunodeficiency virus type 1 NC possesses two zinc finger motifs that are required for specific recognition and packaging of viral genomic RNA. Though essential, zinc fingers and genomic RNA are not required for virion assembly. NC promiscuously associates with cellular RNAs, many of which are incorporated into virions. It has been hypothesized that Gag polymerization and virion assembly are promoted by nonspecific interaction of NC with RNA. Consistent with this model, we found an inverse relationship between the number of NC basic residues replaced with alanine and NC's nonspecific RNA-binding activity, Gag's ability to polymerize in vitro and in vivo, and Gag's capacity to assemble virions. In contrast, mutation of NC's zinc fingers had only minor effects on these properties.
Figures






References
-
- Berkowitz R, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

