cis- and trans-acting elements in flavivirus RNA replication
- PMID: 10708442
- PMCID: PMC111826
- DOI: 10.1128/jvi.74.7.3253-3263.2000
cis- and trans-acting elements in flavivirus RNA replication
Abstract
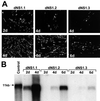
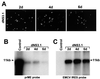
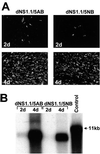
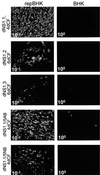
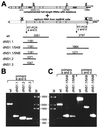
Most of the seven flavivirus nonstructural proteins (NS1 to NS5) encoded in the distal two-thirds of the RNA positive-sense genome are believed to be essential components of RNA replication complexes. To explore the functional relationships of these components in RNA replication, we used trans-complementation analysis of full-length infectious RNAs of Kunjin (KUN) virus with a range of lethal in-frame deletions in the nonstructural coding region, using as helper a repBHK cell line stably producing functional replication complexes from KUN replicon RNA. Recently we showed that replication of KUN RNAs with large carboxy-terminal deletions including the entire RNA polymerase region in the NS5 gene, representing 34 to 75% of the NS5 coding content, could be complemented after transfection into repBHK cells. In this study we have demonstrated that KUN RNAs with deletions of 84 to 97% of the NS1 gene, or of 13 to 63% of the NS3 gene including the entire helicase region, were also complemented in repBHK cells with variable efficiencies. In contrast, KUN RNAs with deletions in any of the other four nonstructural genes NS2A, NS2B, NS4A, and NS4B were not complemented. We have also demonstrated successful trans complementation of KUN RNAs containing either combined double deletions in the NS1 and NS5 genes or triple deletions in the NS1, NS3, and NS5 genes comprising as much as 38% of the entire nonstructural coding content. Based on these and our previous complementation results, we have generated a map of cis- and trans-acting elements in RNA replication for the nonstructural coding region of the flavivirus genome. These results are discussed in the context of our model on formation and composition of the flavivirus replication complex, and we suggest molecular mechanisms by which functions of some defective components of the replication complex can be complemented by their wild-type counterparts expressed from another (helper) RNA molecule.
Figures


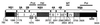
References
-
- Arias C F, Preugschat F, Strauss J H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. - PubMed
-
- Brinkworth R I, Fairlie D P, Leung D, Young P R. Homology model of the dengue 2 virus NS3 protease: putative interactions with both substrate and NS2B cofactor. J Gen Virol. 1999;80:1167–1177. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

