QTL fine mapping by measuring and testing for Hardy-Weinberg and linkage disequilibrium at a series of linked marker loci in extreme samples of populations
- PMID: 10712216
- PMCID: PMC1288140
- DOI: 10.1086/302804
QTL fine mapping by measuring and testing for Hardy-Weinberg and linkage disequilibrium at a series of linked marker loci in extreme samples of populations
Abstract
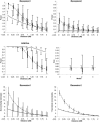
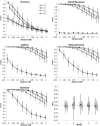
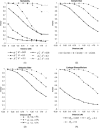
It has recently been demonstrated that fine-scale mapping of a susceptibility locus for a complex disease can be accomplished on the basis of deviations from Hardy-Weinberg (HW) equilibrium at closely linked marker loci among affected individuals. We extend this theory to fine-scale localization of a quantitative-trait locus (QTL) from extreme individuals in populations, by means of HW and linkage-disequilibrium (LD) analyses. QTL mapping and/or linkage analyses can establish a large genomic region ( approximately 30 cM) that contains a QTL. The QTL can be fine mapped by examination of the degree of deviation from HW and LD at a series of closely linked marker loci. The tests can be performed for samples of individuals belonging to either high or low percentiles of the phenotype distribution or for combined samples of these extreme individuals. The statistical properties (the power and the size) of the tests of this fine-mapping approach are investigated and are compared extensively, under various genetic models and parameters for the QTL and marker loci. On the basis of the results, a two-stage procedure that uses extreme samples and different tests (for HW and LD) is suggested for QTL fine mapping. This two-step procedure is economic and powerful and can accurately narrow a genomic region containing a QTL from approximately 30-1 cM, a range that renders physical mapping feasible for identification of the QTL. In addition, the relationship between parameterizations of complex diseases, by means of penetrance, and those of complex quantitative traits, by means of genotypic values, is outlined. This means that many statistical genetic methods developed for searching for susceptibility loci of complex diseases can be directly adopted and/or extended to QTL mapping for quantitative traits.
Figures




Comment in
-
Inflated false-positive rates in Hardy-Weinberg and linkage-equilibrium tests are due to sampling on the basis of rare familial phenotypes in finite populations.Am J Hum Genet. 2000 Jul;67(1):258-9. doi: 10.1086/302964. Am J Hum Genet. 2000. PMID: 10848498 Free PMC article. No abstract available.
-
QTL fine mapping, in extreme samples of finite populations, for complex traits with familial correlation due to polygenes.Am J Hum Genet. 2000 Jul;67(1):259-62. doi: 10.1086/302977. Am J Hum Genet. 2000. PMID: 10848499 Free PMC article. No abstract available.
References
-
- Bengtsson BO, Thomson G (1981) Measuring the strength of association between HLA antigens and diseases. Tissue Antigens 18:356–363 - PubMed
-
- Bennett ST, Lucassen AM, Gough SCL, Powell EE, Undlien DE, Pritchard LE, Merriman ME, et al (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 9:284–292 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

