Molecular basis of variant pseudo-hurler polydystrophy (mucolipidosis IIIC)
- PMID: 10712439
- PMCID: PMC289169
- DOI: 10.1172/JCI5826
Molecular basis of variant pseudo-hurler polydystrophy (mucolipidosis IIIC)
Abstract
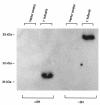
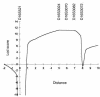
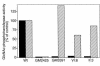
Mucolipidosis IIIC, or variant pseudo-Hurler polydystrophy, is an autosomal recessive disease of lysosomal hydrolase trafficking. Unlike the related diseases, mucolipidosis II and IIIA, the enzyme affected in mucolipidosis IIIC (N-Acetylglucosamine-1-phosphotransferase [GlcNAc-phosphotransferase]) retains full transferase activity on synthetic substrates but lacks activity on lysosomal hydrolases. Bovine GlcNAc-phosphotransferase has recently been isolated as a multisubunit enzyme with the subunit structure alpha(2)beta(2)gamma(2). We cloned the cDNA for the human gamma-subunit and localized its gene to chromosome 16p. We also showed, in a large multiplex Druze family that exhibits this disorder, that MLIIIC also maps to this chromosomal region. Sequence analysis of the gamma-subunit cDNA in patients from 3 families identified a frameshift mutation, in codon 167 of the gamma subunit, that segregated with the disease, indicating MLIIIC results from mutations in the phosphotransferase gamma-subunit gene. This is to our knowledge the first description of the molecular basis for a human mucolipidosis and suggests that the gamma subunit functions in lysosomal hydrolase recognition.
Figures



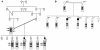
Comment in
-
The missing link in lysosomal enzyme targeting.J Clin Invest. 2000 Mar;105(5):563-4. doi: 10.1172/JCI9479. J Clin Invest. 2000. PMID: 10712426 Free PMC article. No abstract available.
References
-
- Leroy JG, DeMars RI. Mutant enzymatic and cytological phenotypes in cultured human fibroblasts. Science. 1967;157:804–806. - PubMed
-
- Hickman S, Neufeld EF. A hypothesis for I-cell disease: defective hydrolases that do not enter lysosomes. Biochem Biophys Res Commun. 1972;4:992–999. - PubMed
-
- Hickman S, Shapiro LJ, Neufeld EF. A recognition marker required for uptake of a lysosomal enzyme by cultured fibroblasts. Biochem Biophys Res Commun. 1974;1:55–61. - PubMed
-
- Kornfeld S. Trafficking of lysosomal enzymes. FASEB J. 1987;1:462–468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

