Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex
- PMID: 10712509
- PMCID: PMC14820
- DOI: 10.1091/mbc.11.3.915
Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex
Abstract
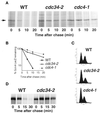
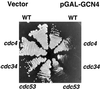
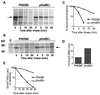
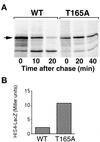
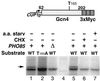
Gcn4, a yeast transcriptional activator that promotes the expression of amino acid and purine biosynthesis genes, is rapidly degraded in rich medium. Here we report that SCF(CDC4), a recently characterized protein complex that acts in conjunction with the ubiquitin-conjugating enzyme Cdc34 to degrade cell cycle regulators, is also necessary for the degradation of the transcription factor Gcn4. Degradation of Gcn4 occurs throughout the cell cycle, whereas degradation of the known cell cycle substrates of Cdc34/SCF(CDC4) is cell cycle regulated. Gcn4 ubiquitination and degradation are regulated by starvation for amino acids, whereas the degradation of the cell cycle substrates of Cdc34/SCF(CDC4) is unaffected by starvation. We further show that unlike the cell cycle substrates of Cdc34/SCF(CDC4), which require phosphorylation by the kinase Cdc28, Gcn4 degradation requires the kinase Pho85. We identify the critical target site of Pho85 on Gcn4; a mutation of this site stabilizes the protein. A specific Pho85-Pcl complex that is able to phosphorylate Gcn4 on that site is inactive under conditions under which Gcn4 is stable. Thus, Cdc34/SCF(CDC4) activity is constitutive, and regulation of the stability of its various substrates occurs at the level of their phosphorylation.
Figures





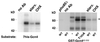

References
-
- Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. - PubMed
-
- Arndt KT, Styles CA, Fink GR. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. - PubMed
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989.
-
- Bai C, Sen P, Hoffman K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

