The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein
- PMID: 10713161
- PMCID: PMC85410
- DOI: 10.1128/MCB.20.7.2367-2377.2000
The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein
Abstract
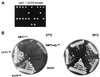
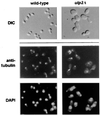
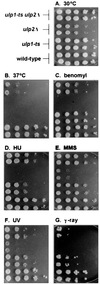
Yeast Smt3 and its vertebrate homolog SUMO-1 are ubiquitin-like proteins (Ubls) that are reversibly ligated to other proteins. Like SMT3, SMT4 was first isolated as a high-copy-number suppressor of a defective centromere-binding protein. We show here that SMT4 encodes an Smt3-deconjugating enzyme, Ulp2. In cells lacking Ulp2, specific Smt3-protein conjugates accumulate, and the conjugate pattern is distinct from that observed in a ulp1(ts) strain, which is defective for a distantly related Smt3-specific protease, Ulp1. The ulp2Delta mutant exhibits a pleiotropic phenotype that includes temperature-sensitive growth, abnormal cell morphology, decreased plasmid and chromosome stability, and a severe sporulation defect. The mutant is also hypersensitive to DNA-damaging agents, hydroxyurea, and benomyl. Although cell cycle checkpoint arrest in response to DNA damage, replication inhibition, or spindle defects occurs with normal kinetics, recovery from arrest is impaired. Surprisingly, either introduction of a ulp1(ts) mutation or overproduction of catalytically inactive Ulp1 can substantially overcome the ulp2Delta defects. Inactivation of Ulp2 also suppresses several ulp1(ts) defects, and the double mutant accumulates far fewer Smt3-protein conjugates than either single mutant. Our data suggest the existence of a feedback mechanism that limits Smt3-protein ligation when Smt3 deconjugation by both Ulp1 and Ulp2 is compromised, allowing a partial recovery of cell function.
Figures


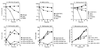
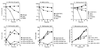

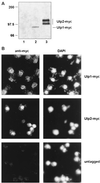

References
-
- al-Khodairy F, Enoch T, Hagan I M, Carr A M. The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J Cell Sci. 1995;108:475–486. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989.
-
- Bayer P, Arndt A, Metzger S, Mahajan R, Mechior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. - PubMed
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. - PubMed
-
- Desterro J M, Rodriguez M S, Kemp G D, Hay R T. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274:10618–10624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
