Unambiguous demonstration of triple-helix-directed gene modification
- PMID: 10716704
- PMCID: PMC16196
- DOI: 10.1073/pnas.97.7.3084
Unambiguous demonstration of triple-helix-directed gene modification
Abstract
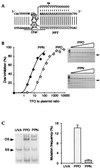
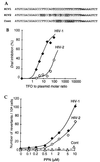
Triple-helix-forming oligonucleotides (TFOs), which can potentially modify target genes irreversibly, represent promising tools for antiviral therapies. However, their effectiveness on endogenous genes has yet to be unambiguously demonstrated. To monitor endogenous gene modification by TFOs in a yeast model, we inactivated an auxotrophic marker gene by inserting target sequences of interest into its coding region. The genetically engineered yeast cells then were treated with psoralen-linked TFOs followed by UV irradiation, thus generating highly mutagenic covalent crosslinks at the target site whose repair could restore gene function; the number of revertants and spectrum of mutations generated were quantified. Results showed that a phosphoramidate TFO indeed reaches its target sequence, forms crosslinks, and generates mutations at the expected site via a triplex-mediated mechanism: (i) under identical conditions, no mutations were generated by the same TFO at two other loci in the target strain, nor in an isogenic control strain carrying a modified target sequence incapable of supporting triple-helix formation; (ii) for a given target sequence, whether the triplex was formed in vivo on an endogenous gene or in vitro on an exogenous plasmid, the nature of the mutations generated was identical, and consistent with the repair of a psoralen crosslink at the target site. Although the mutation efficiency was probably too low for therapeutic applications, our results confirm the validity of the triple-helix approach and provide a means of evaluating the effectiveness of new chemically modified TFOs and analogs.
Figures


References
-
- Hélène C, Giovannangeli C, Guieysse-Peugeot A L, Praseuth D. Ciba Found Symp. 1997;209:94–102. - PubMed
-
- Malvy C, Harel-Bellan A, Pritchard L L, editors. Triple Helix Forming Oligonucleotides. Boston: Kluwer; 1999.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

