Fission yeast switches mating type by a replication-recombination coupled process
- PMID: 10716938
- PMCID: PMC305679
- DOI: 10.1093/emboj/19.6.1389
Fission yeast switches mating type by a replication-recombination coupled process
Abstract
Fission yeast exhibits a homothallic life cycle, in which the mating type of the cell mitotically alternates in a highly regulated fashion. Pedigree analysis of dividing cells has shown that only one of the two sister cells switches mating type. It was shown recently that a site- and strand-specific DNA modification at the mat1 locus precedes mating-type switching. By tracking the fate of mat1 DNA throughout the cell cycle with a PCR assay, we identified a novel DNA intermediate of mating-type switching in S-phase. The time and rate of appearance and disappearance of this DNA intermediate are consistent with a model in which mating-type switching occurs through a replication-recombination coupled pathway. Such a process provides experimental evidence in support of a copy choice recombination model in Schizosaccharomyces pombe mating-type switching and is reminiscent of the sister chromatid recombination used to complete replication in the presence of certain types of DNA damage.
Figures

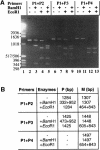
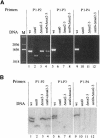
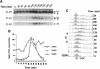


References
-
- Bardwell A.J., Bardwell, L., Tomkinson, A.E. and Friedberg, E.C. (1994) Specific cleavage of model recombination and repair intermediates by the yeast Rad1–Rad10 DNA endonuclease. Science, 265, 2082–2085. - PubMed
-
- Beach D.H. (1983) Cell type switching by DNA transposition in fission yeast. Nature, 305, 682–688.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

