The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development
- PMID: 10716941
- PMCID: PMC316425
The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development
Abstract
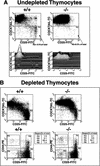
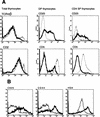
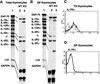
SATB1 is expressed primarily in thymocytes and can act as a transcriptional repressor. SATB1 binds in vivo to the matrix attachment regions (MARs) of DNA, which are implicated in the loop domain organization of chromatin. The role of MAR-binding proteins in specific cell lineages is unknown. We generated SATB1-null mice to determine how SATB1 functions in the T-cell lineage. SATB1-null mice are small in size, have disproportionately small thymi and spleens, and die at 3 weeks of age. At the cellular level, multiple defects in T-cell development were observed. Immature CD3(-)CD4(-)CD8(-) triple negative (TN) thymocytes were greatly reduced in number, and thymocyte development was blocked mainly at the DP stage. The few peripheral CD4(+) single positive (SP) cells underwent apoptosis and failed to proliferate in response to activating stimuli. At the molecular level, among 589 genes examined, at least 2% of genes including a proto-oncogene, cytokine receptor genes, and apoptosis-related genes were derepressed at inappropriate stages of T-cell development in SATB1-null mice. For example, IL-2Ralpha and IL-7Ralpha genes were ectopically transcribed in CD4(+)CD8(+) double positive (DP) thymocytes. SATB1 appears to orchestrate the temporal and spatial expression of genes during T-cell development, thereby ensuring the proper development of this lineage. Our data provide the first evidence that MAR-binding proteins can act as global regulators of cell function in specific cell lineages.
Figures





References
-
- Adams MD, Kerlavage AR, Fields C, Venter JC. 3,400 new expressed sequence tags identify diversity of transcripts in human brain. Nat Genet. 1993;4:256–267. - PubMed
-
- Agata Y, Kawasaki A, Nishimura H, Ishida Y, Tsubata T, Yagita H, Honjo T. Expression of the PD-1 antigen on the surface of stimulated mouse T and B lymphocytes. Int Immunol. 1996;8:765–772. - PubMed
-
- Banan M, Rojas IC, Lee W-H, King HL, Harriss JV, Kobayashi R, Webb CF, Gottlieb PD. Interaction of the nuclear matrix-associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem. 1997;272:18440–18452. - PubMed
-
- Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
