The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure
- PMID: 10716989
- PMCID: PMC15968
- DOI: 10.1073/pnas.97.6.2562
The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure
Abstract
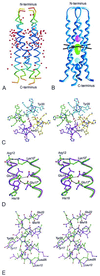
The question of whether a protein whose natural sequence is inverted adopts a stable fold is still under debate. We have determined the 2. 1-A crystal structure of the retro-GCN4 leucine zipper. In contrast to the two-stranded helical coiled-coil GCN4 leucine zipper, the retro-leucine zipper formed a very stable, parallel four-helix bundle, which now lends itself to further structural and functional studies.
Figures

References
-
- Anfinsen C B. Science. 1973;181:223–230. - PubMed
-
- Guptasarma P. FEBS Lett. 1992;310:205–210. - PubMed
-
- Olszewski K A, Kolinski A, Skolnick J. Protein Eng. 1996;9:5–14. - PubMed
-
- Witte K, Skolnick J, Wong C-H. J Am Chem Soc. 1998;120:13042–13046.
-
- Beck K, Brodsky B. J Struct Biol. 1998;122:17–29. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

