Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis
- PMID: 10716996
- PMCID: PMC15989
- DOI: 10.1073/pnas.97.6.2680
Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis
Abstract
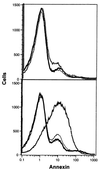
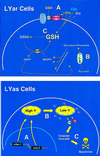
Multigenic programs controlling susceptibility to apoptosis in response to ionizing radiation have not yet been defined. Here, using DNA microarrays, we show gene expression patterns in an apoptosis-sensitive and apoptosis-resistant murine B cell lymphoma model system both before and after irradiation. From the 11,000 genes interrogated by the arrays, two major patterns emerged. First, before radiation exposure the radioresistant LYar cells expressed significantly greater levels of message for several genes involved in regulating intracellular redox potential. Compared with LYas cells, LYar cells express 20- to 50-fold more mRNA for the tetraspanin CD53 and for fructose-1,6-bisphosphatase. Expression of both of these genes can lead to the increase of total cellular glutathione, which is the principle intracellular antioxidant and has been shown to inhibit many forms of apoptosis. A second pattern emerged after radiation, when the apoptosis-sensitive LYas cells induced rapid expression of a unique cluster of genes characterized by their involvement in mitochondrial electron transport. Some of these genes have been previously recognized as proapoptotic; however others, such as uncoupling protein 2, were not previously known to be apoptotic regulatory proteins. From these observations we propose that a multigenic program for sensitivity to apoptosis involves induction of transcripts for genes participating in mitochondrial uncoupling and loss of membrane potential. This program triggers mitochondrial release of apoptogenic factors and induces the "caspase cascade." Conversely, cells resistant to apoptosis down-regulate these biochemical pathways, while activating pathways for establishment and maintenance of high intracellular redox potential by means of elevated glutathione.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

