Nucleotide sequence of the bla(RTG-2) (CARB-5) gene and phylogeny of a new group of carbenicillinases
- PMID: 10722515
- PMCID: PMC89816
- DOI: 10.1128/AAC.44.4.1070-1074.2000
Nucleotide sequence of the bla(RTG-2) (CARB-5) gene and phylogeny of a new group of carbenicillinases
Abstract
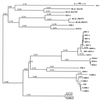
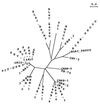
We determined the nucleotide sequence of the bla gene for the Acinetobacter calcoaceticus beta-lactamase previously described as CARB-5. Alignment of the deduced amino acid sequence with those of known beta-lactamases revealed that CARB-5 possesses an RTG triad in box VII, as described for the Proteus mirabilis GN79 enzyme, instead of the RSG consensus characteristic of the other carbenicillinases. Phylogenetic studies showed that these RTG enzymes constitute a new, separate group, possibly ancestors of the carbenicillinase family.
Figures


References
-
- Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond Biol. 1980;289:321–331. - PubMed
-
- Boissinot M, Levesque R C. Nucleotide sequence of the PSE-4 carbenicillinase gene and correlations with the Staphylococcus aureus PC1 β-lactamase crystal structure. J Biol Chem. 1990;265:1225–1230. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

