Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila
- PMID: 10722574
- PMCID: PMC97358
- DOI: 10.1128/IAI.68.4.1855-1863.2000
Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila
Abstract
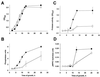
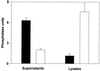
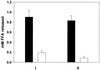
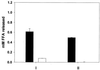
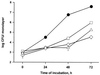
Legionella pneumophila, the agent of Legionnaires' disease, is an intracellular pathogen of protozoa and macrophages. Previously, we had determined that the Legionella pilD gene is involved in type IV pilus biogenesis, type II protein secretion, intracellular infection, and virulence. Since the loss of pili and a protease do not account for the infection defect exhibited by a pilD-deficient strain, we sought to define other secreted proteins absent in the mutant. Based upon the release of p-nitrophenol (pNP) from p-nitrophenyl phosphate, acid phosphatase activity was detected in wild-type but not in pilD mutant supernatants. Mutant supernatants also did not release either pNP from p-nitrophenyl caprylate and palmitate or free fatty acid from 1-monopalmitoylglycerol, suggesting that they lack a lipase-like activity. However, since wild-type samples failed to release free fatty acids from 1,2-dipalmitoylglycerol or to cleave a triglyceride derivative, this secreted activity should be viewed as an esterase-monoacylglycerol lipase. The mutant supernatants were defective for both release of free fatty acids from phosphatidylcholine and degradation of RNA, indicating that PilD-negative bacteria lack a secreted phospholipase A (PLA) and nuclease. Finally, wild-type but not mutant supernatants liberated pNP from p-nitrophenylphosphorylcholine (pNPPC). Characterization of a new set of mutants defective for pNPPC-hydrolysis indicated that this wild-type activity is due to a novel enzyme, as opposed to a PLC or another known enzyme. Some, but not all, of these mutants were greatly impaired for intracellular infection, suggesting that a second regulator or processor of the pNPPC hydrolase is critical for L. pneumophila virulence.
Figures
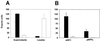



References
-
- Alm R A, Hallinan J P, Watson A A, Mattick J S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. - PubMed
-
- Babich H, Borenfreund E, Stern A. Comparative cytotoxicities of selected minor dietary non-nutrients with chemopreventive properties. Cancer Lett. 1993;73:127–133. - PubMed
-
- Baine W B. Cytolytic and phospholipase C activity in Legionella species. J Gen Microbiol. 1985;131:1383–1391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

